RetrogeneDB ID: | retro_mmus_158 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 5:7179364..7180699(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000095159 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tubb4b | ||
| Ensembl ID: | ENSMUSG00000036752 | ||
| Aliases: | Tubb4b, 4930542G03Rik, Tubb2c, Tubb2c1 | ||
| Description: | tubulin, beta 4B class IVB [Source:MGI Symbol;Acc:MGI:1915472] |

Retrocopy-Parental alignment summary:
>retro_mmus_158
ATGAGGGAGATCGTGCACCTGCAGGCTGGGCAGTGCGGCAACCAGATTGGCGCCAAGTTCTGGGAGGTAATCAGTGACGA
GCATGGCATTGATACCACTGGCACTTACCACGGAGATAGCGACCTCCAGCTGGAGCGCATCAATGTGTATTACAACGAAG
CCACCGGTGGCAAGTATGTGCCCCGCGCCGTGCTCGTGGACTTGGAGCCCTGCACCATGGACTCGGTTCGTTCAGGGCCC
TTCGGGCAGATCTTCAGACCTGATAACTTCGTCTTTGGTCAGAGTGGGGCTGGGAACAACTGGGCCAAGGGGCACTACAC
AGAAGGTGCAGAGCTGGTTGACTCGGTGTTGGACGTTGTGAGAAAGGAAGCTGAGAGCTGTGACTGCCTGCAGGGCTTCC
AGCTGACCCACTCCCTGGGTGGGGGGACTGGGTCTGGGATGGGTACCCTCATCAGCAAGATCCGGGAGGAGTACCCAGAC
AGAATCATGAACACCTTCAGTGTGGTACCTTCCCCCAAGGTGTCGGACACAGTGGTTGAGCCCTACAATGCCACCCTTTC
AGTCCATCAGCTGGTTGAAAACACAGATGAGACCTATTGTATTGATGATGAAGCACTCTATGACATCTGCTTCAGAACCC
TAAAGCTGACCACACCCACTTACGGTGACCTGAACCATCTAGTGTCCGCCACCATGAGTGGGGTAACCACTTGCCTGCGA
TTCCCTGGCCAGCTAAATGCTGACCTGCGGAAACTGGCTGTAAATATGGTGCCCTTCCCTCGCCTGCACTTCTTCATGCC
TGGCTTTGCCCCCTTGACCAGCCGGGGAAGCCAGCAGTACCGTGCCCTGACAGTTCCTGAGCTCACCCAGCAGATGTTTG
ATGCCAAGAACATGATGGCTGCCTGTGATCCAAGACATGGGCGCTACTTGACTGTGGCTGCCGTGTTCAGGGGCCGCATG
TCTATGAAGGAGGTGGACGAACAGATGCTTAATGTCCAAAACAAGAACAGCAGCTACTTTGTTGAGTGGATCCCCAACAA
TGTGAAGACAGCTGTCTGTGACATTCCACCTCGGGGCCTGAAAATGTCCACCACCTTCATTGGCAACAGCACCGCTATCC
AGGAGCTGTTCAAACGCATCTCAGAGCAGTTCACAGCCATGTTCCCACGCAAGACCTTCCTACATTGGTACACGGGTGAG
GGCATGGATGAGATGGAGTTCACTGAGGCTGAGAGCAACATGAACGACCTGGTGTCCGAGTACCAGCAGTACCAGGATGC
TACAGCTGAGGAAGAGGGAGAGTATGAGGAGGAGGCTGAGGAGGAGGTGGCTTAG
ATGAGGGAGATCGTGCACCTGCAGGCTGGGCAGTGCGGCAACCAGATTGGCGCCAAGTTCTGGGAGGTAATCAGTGACGA
GCATGGCATTGATACCACTGGCACTTACCACGGAGATAGCGACCTCCAGCTGGAGCGCATCAATGTGTATTACAACGAAG
CCACCGGTGGCAAGTATGTGCCCCGCGCCGTGCTCGTGGACTTGGAGCCCTGCACCATGGACTCGGTTCGTTCAGGGCCC
TTCGGGCAGATCTTCAGACCTGATAACTTCGTCTTTGGTCAGAGTGGGGCTGGGAACAACTGGGCCAAGGGGCACTACAC
AGAAGGTGCAGAGCTGGTTGACTCGGTGTTGGACGTTGTGAGAAAGGAAGCTGAGAGCTGTGACTGCCTGCAGGGCTTCC
AGCTGACCCACTCCCTGGGTGGGGGGACTGGGTCTGGGATGGGTACCCTCATCAGCAAGATCCGGGAGGAGTACCCAGAC
AGAATCATGAACACCTTCAGTGTGGTACCTTCCCCCAAGGTGTCGGACACAGTGGTTGAGCCCTACAATGCCACCCTTTC
AGTCCATCAGCTGGTTGAAAACACAGATGAGACCTATTGTATTGATGATGAAGCACTCTATGACATCTGCTTCAGAACCC
TAAAGCTGACCACACCCACTTACGGTGACCTGAACCATCTAGTGTCCGCCACCATGAGTGGGGTAACCACTTGCCTGCGA
TTCCCTGGCCAGCTAAATGCTGACCTGCGGAAACTGGCTGTAAATATGGTGCCCTTCCCTCGCCTGCACTTCTTCATGCC
TGGCTTTGCCCCCTTGACCAGCCGGGGAAGCCAGCAGTACCGTGCCCTGACAGTTCCTGAGCTCACCCAGCAGATGTTTG
ATGCCAAGAACATGATGGCTGCCTGTGATCCAAGACATGGGCGCTACTTGACTGTGGCTGCCGTGTTCAGGGGCCGCATG
TCTATGAAGGAGGTGGACGAACAGATGCTTAATGTCCAAAACAAGAACAGCAGCTACTTTGTTGAGTGGATCCCCAACAA
TGTGAAGACAGCTGTCTGTGACATTCCACCTCGGGGCCTGAAAATGTCCACCACCTTCATTGGCAACAGCACCGCTATCC
AGGAGCTGTTCAAACGCATCTCAGAGCAGTTCACAGCCATGTTCCCACGCAAGACCTTCCTACATTGGTACACGGGTGAG
GGCATGGATGAGATGGAGTTCACTGAGGCTGAGAGCAACATGAACGACCTGGTGTCCGAGTACCAGCAGTACCAGGATGC
TACAGCTGAGGAAGAGGGAGAGTATGAGGAGGAGGCTGAGGAGGAGGTGGCTTAG
ORF - retro_mmus_158 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 98.2 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MREIVHLQAGQCGNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEP |
| MREIVHLQAGQCGNQIGAKFWEVISDEHGID.TGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEP | |
| Retrocopy | MREIVHLQAGQCGNQIGAKFWEVISDEHGIDTTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEP |
| Parental | GTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQGFQLTHSLG |
| .TMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQGFQLTHSLG | |
| Retrocopy | CTMDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQGFQLTHSLG |
| Parental | GGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCIDNEALYDI |
| GGTGSGMGTL.ISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCID.EALYDI | |
| Retrocopy | GGTGSGMGTL-ISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQLVENTDETYCIDDEALYDI |
| Parental | CFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQ |
| CFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQ | |
| Retrocopy | CFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRKLAVNMVPFPRLHFFMPGFAPLTSRGSQQ |
| Parental | YRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVK |
| YRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVK | |
| Retrocopy | YRALTVPELTQQMFDAKNMMAACDPRHGRYLTVAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVK |
| Parental | TAVCDIPPRGLKMSATFIGNSTAIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVS |
| TAVCDIPPRGLKMS.TFIGNSTAIQELFKRISEQFTAMF.RK.FLHWYTGEGMDEMEFTEAESNMNDLVS | |
| Retrocopy | TAVCDIPPRGLKMSTTFIGNSTAIQELFKRISEQFTAMFPRKTFLHWYTGEGMDEMEFTEAESNMNDLVS |
| Parental | EYQQYQDATAEEEGEFEEEAEEEVA |
| EYQQYQDATAEEEGE.EEEAEEEVA | |
| Retrocopy | EYQQYQDATAEEEGEYEEEAEEEVA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .84 RPM | 107 .19 RPM |
| SRP007412_cerebellum | 1 .56 RPM | 92 .70 RPM |
| SRP007412_heart | 0 .16 RPM | 76 .24 RPM |
| SRP007412_kidney | 1 .44 RPM | 65 .66 RPM |
| SRP007412_liver | 0 .41 RPM | 31 .05 RPM |
| SRP007412_testis | 33 .16 RPM | 443 .58 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_158 was not detected
No EST(s) were mapped for retro_mmus_158 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_115115 | 196 libraries | 306 libraries | 237 libraries | 179 libraries | 154 libraries |
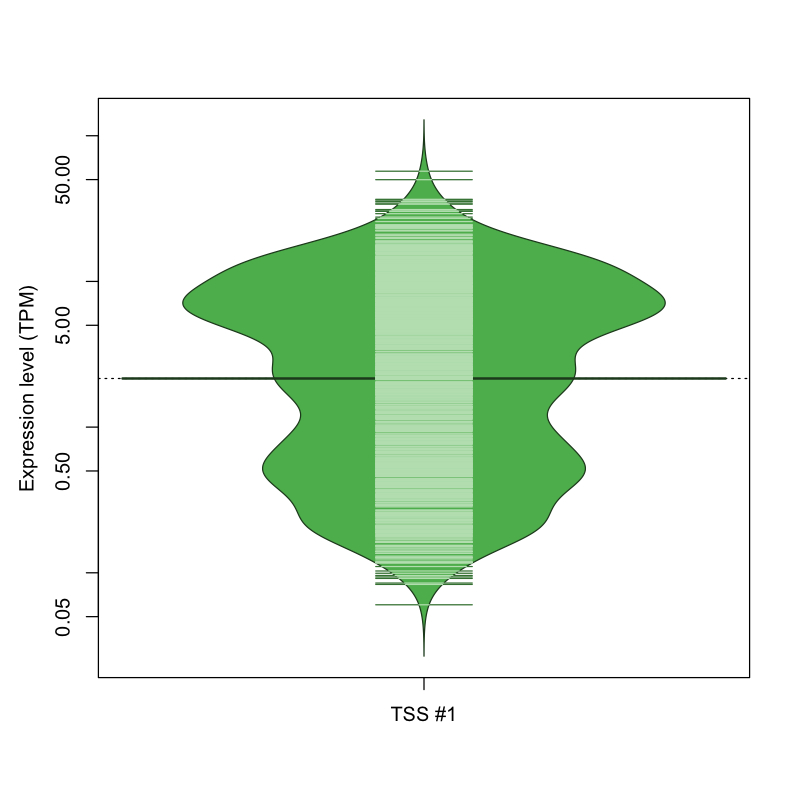
The graphical summary, for retro_mmus_158 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_158 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_158 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 10 parental genes, and 16 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000016380 | 2 retrocopies | |
| Cavia porcellus | ENSCPOG00000011966 | 1 retrocopy | |
| Microcebus murinus | ENSMICG00000010939 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000012164 | 2 retrocopies | |
| Monodelphis domestica | ENSMODG00000017213 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000016255 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000036752 | 2 retrocopies |
retro_mmus_158 , retro_mmus_422,
|
| Otolemur garnettii | ENSOGAG00000005927 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000023373 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000010170 | 2 retrocopies |