RetrogeneDB ID: | retro_mmus_294 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:9450655..9451743(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Hibch | ||
| Ensembl ID: | ENSMUSG00000041426 | ||
| Aliases: | Hibch, 2610509I15Rik, AI648812 | ||
| Description: | 3-hydroxyisobutyryl-Coenzyme A hydrolase [Source:MGI Symbol;Acc:MGI:1923792] |

Retrocopy-Parental alignment summary:
>retro_mmus_294
AGTGTTATTCTGCAACAACTGAGAATGTCCATGCACACAGAAGCTGCAGAAGTGCTACTGGAGAGGAGAGGCTGTGCAGG
GGTCATAACACTCAACAGACTCAATGCACTGAGTCTGAATATGATCCAGCAGATCTATCCACAGCTAAAGAAGTGGAAAC
AAGACCTTGACACATTCCTGATCATCATAAAGGGAGCCTGAGGAAAGGCCTTTTGTGCTGGAGTGGACATCAAAGCACTC
TCAGAAGCTAAAAAAGCGAGACAGAATTGGACTCATGATGTATTCAGAGAAGAATACATCCTGAACAATGCAATTGCATC
TTGTCAGAAACCATATGTTGCCCTTATTGATGGAATCACCATGTTTGGGGAAGTTGGTCTTTCAGTTCATGGTCATTTCC
GAGTGGCTACAGAAAGGTCTCTCTTTGCTATGCCAGAAACAGGAATAGGACTATTTCCTGATGTGGGTAGAGGTTATTTC
CTGCCAACATTTCAAGGAAAACTGGGTTACTTCCTTGCACTGACAGGACACAGATTAAAAGGAAGAGATGTGCACAGAGC
GGGAATTGACACACACTTTGTAGATTCAGAAAAGTTGCATGTGTTGGAGGAGGAGCTGTTAGCCTTAAACTCACCTCCAG
CTGAGGATGTTGCAGGTGTCTTAGAAAACTATCATGCCAAGTCCAAGATGGATCAAGACAAATCTTTATATTTGAGGAAC
ACATGGGCAAAATAAACAGTTGTTTTTCAGCCAATACCATGGAACAGATAATTGAAAACTTATGGCAAAATGGATCACCT
TTTGCCATAGAATAGATGAAGGTAATTAATAAAATGTCTCTGACATCACTAAAGATCACACGAAGGCAGCTCATGGAGGG
TTCTTCAAAGACCTTGCAAGAGGTACTGACCATGGAATATCGGTTAACTCAAGCATGTATGGAAGGTTATAACTTTCATG
AAGGTGTTAGAGTTGTTATCATTGACAAAGACCAGACTTCAACATGGAAACCAGCCAGTCTAAAAGATTTTACTGATAAA
GATTTGAATAGTTATTTTAAATCTCTGGGAAGCACTGATTTGAAATTC
AGTGTTATTCTGCAACAACTGAGAATGTCCATGCACACAGAAGCTGCAGAAGTGCTACTGGAGAGGAGAGGCTGTGCAGG
GGTCATAACACTCAACAGACTCAATGCACTGAGTCTGAATATGATCCAGCAGATCTATCCACAGCTAAAGAAGTGGAAAC
AAGACCTTGACACATTCCTGATCATCATAAAGGGAGCCTGAGGAAAGGCCTTTTGTGCTGGAGTGGACATCAAAGCACTC
TCAGAAGCTAAAAAAGCGAGACAGAATTGGACTCATGATGTATTCAGAGAAGAATACATCCTGAACAATGCAATTGCATC
TTGTCAGAAACCATATGTTGCCCTTATTGATGGAATCACCATGTTTGGGGAAGTTGGTCTTTCAGTTCATGGTCATTTCC
GAGTGGCTACAGAAAGGTCTCTCTTTGCTATGCCAGAAACAGGAATAGGACTATTTCCTGATGTGGGTAGAGGTTATTTC
CTGCCAACATTTCAAGGAAAACTGGGTTACTTCCTTGCACTGACAGGACACAGATTAAAAGGAAGAGATGTGCACAGAGC
GGGAATTGACACACACTTTGTAGATTCAGAAAAGTTGCATGTGTTGGAGGAGGAGCTGTTAGCCTTAAACTCACCTCCAG
CTGAGGATGTTGCAGGTGTCTTAGAAAACTATCATGCCAAGTCCAAGATGGATCAAGACAAATCTTTATATTTGAGGAAC
ACATGGGCAAAATAAACAGTTGTTTTTCAGCCAATACCATGGAACAGATAATTGAAAACTTATGGCAAAATGGATCACCT
TTTGCCATAGAATAGATGAAGGTAATTAATAAAATGTCTCTGACATCACTAAAGATCACACGAAGGCAGCTCATGGAGGG
TTCTTCAAAGACCTTGCAAGAGGTACTGACCATGGAATATCGGTTAACTCAAGCATGTATGGAAGGTTATAACTTTCATG
AAGGTGTTAGAGTTGTTATCATTGACAAAGACCAGACTTCAACATGGAAACCAGCCAGTCTAAAAGATTTTACTGATAAA
GATTTGAATAGTTATTTTAAATCTCTGGGAAGCACTGATTTGAAATTC
ORF - retro_mmus_294 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 87.19 % |
| Parental protein coverage: | 95.06 % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | SVILQHLRMSMHTEAAEVLLERRGCGGVITLNRPKFLNALSLNMIRQIYPQLKTWEQDPDTFLIIIKGAG |
| SVILQ.LRMSMHTEAAEVLLERRGC.GVITLNR...LNALSLNMI.QIYPQLK.W.QD.DTFLIIIKGA. | |
| Retrocopy | SVILQQLRMSMHTEAAEVLLERRGCAGVITLNR---LNALSLNMIQQIYPQLKKWKQDLDTFLIIIKGA* |
| Parental | GKAFCAGGDIKALSEAKKARQNLTQDLFREEYILNNAIASCQKPYVALIDGITMGGGVGLSVHGQFRVAT |
| GKAFCAG.DIKALSEAKKARQN.T.D.FREEYILNNAIASCQKPYVALIDGITM.G.VGLSVHG.FRVAT | |
| Retrocopy | GKAFCAGVDIKALSEAKKARQNWTHDVFREEYILNNAIASCQKPYVALIDGITMFGEVGLSVHGHFRVAT |
| Parental | ERSLFAMPETGIGLFPDVGGGYFLPRLQGKLGYFLALTGYRLKGRDVHRAGIATHFVDSEKLRVLEEELL |
| ERSLFAMPETGIGLFPDVG.GYFLP..QGKLGYFLALTG.RLKGRDVHRAGI.THFVDSEKL.VLEEELL | |
| Retrocopy | ERSLFAMPETGIGLFPDVGRGYFLPTFQGKLGYFLALTGHRLKGRDVHRAGIDTHFVDSEKLHVLEEELL |
| Parental | ALKSPSAEDVAGVLESYHAKSKMDQDKSI-IFEEHMDKINSCFSANTVEQIIENLRQDGSPFAIEQMKVI |
| AL.SP.AEDVAGVLE.YHAKSKMDQDKS..IFEEHM.KINSCFSANT.EQIIENL.Q.GSPFAIE.MKVI | |
| Retrocopy | ALNSPPAEDVAGVLENYHAKSKMDQDKSL<IFEEHMGKINSCFSANTMEQIIENLWQNGSPFAIE*MKVI |
| Parental | NKMSPTSLKITLRQLMEGSSKTLQEVLIMEYRITQACMEGHDFHEGVRAVLIDKDQTPKWKPANLKDVTD |
| NKMS.TSLKIT.RQLMEGSSKTLQEVL.MEYR.TQACMEG..FHEGVR.V.IDKDQT..WKPA.LKD.TD | |
| Retrocopy | NKMSLTSLKITRRQLMEGSSKTLQEVLTMEYRLTQACMEGYNFHEGVRVVIIDKDQTSTWKPASLKDFTD |
| Parental | EDLNSYFKSLGSSDLKF |
| .DLNSYFKSLGS.DLKF | |
| Retrocopy | KDLNSYFKSLGSTDLKF |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 10 .54 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 12 .90 RPM |
| SRP007412_heart | 0 .00 RPM | 49 .19 RPM |
| SRP007412_kidney | 0 .02 RPM | 44 .95 RPM |
| SRP007412_liver | 0 .00 RPM | 53 .63 RPM |
| SRP007412_testis | 0 .05 RPM | 4 .85 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_294 was not detected
No EST(s) were mapped for retro_mmus_294 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_77718 | 862 libraries | 192 libraries | 16 libraries | 1 library | 1 library |
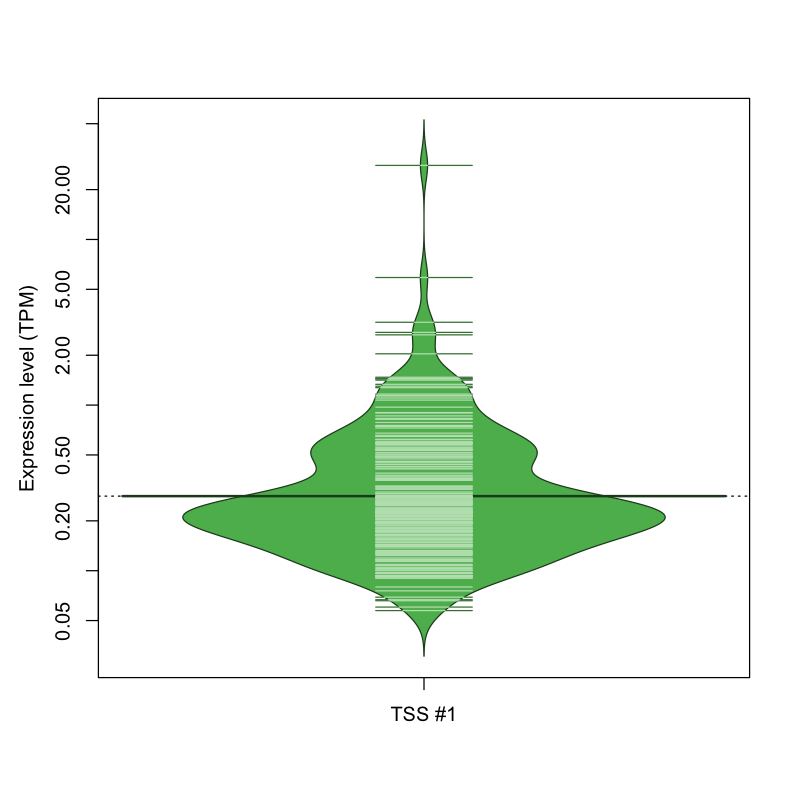
The graphical summary, for retro_mmus_294 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_294 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_294 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 9 parental genes, and 10 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Felis catus | ENSFCAG00000003341 | 1 retrocopy | |
| Homo sapiens | ENSG00000198130 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000004988 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000007283 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000041426 | 1 retrocopy |
retro_mmus_294 ,
|
| Nomascus leucogenys | ENSNLEG00000006646 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000013021 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000012741 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000004132 | 1 retrocopy |