RetrogeneDB ID: | retro_mmus_3520 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:128261656..128262843(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000081739 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Mdm4 | ||
| Ensembl ID: | ENSMUSG00000054387 | ||
| Aliases: | Mdm4, 4933417N07Rik, AA414968, AL023055, AU018793, AU021806, C85810, Mdmx | ||
| Description: | transformed mouse 3T3 cell double minute 4 [Source:MGI Symbol;Acc:MGI:107934] |

Retrocopy-Parental alignment summary:
>retro_mmus_3520
GGATGTCAGAACTTTCTGTGAAAGATCCAAGCCCTCTCTATGACGTGGCTAAGAAAGAATCTTCTATTCACACAGATGCT
GCTCAGACTCTCGCTCTCACACAGGATCACGCTATGGATTTCCCAAGTCAAGACCAACTGAAGCACGGCGCAACAGAGCA
CTCCGATCCCAGAAAAACAACTGAAGAAGAGGATACTCACACACTGCCTACCTCACCACGTAGATGCAGAGAGTCCAAAA
CAGATGAAGACTTGATAGAACATGTATCTCAAGATGAGACATCTAGGCTTGAACTTGATTTTGAGGAGTGGGACGTTGCT
GGCCTGCCTTGGTGGTTTCTAGGGAATTTGAGAAACGACGATATTCCTAAAAGTAATGGCTCAACTGATTTACAGACAAA
TCAGGATATAGGTACTGCAATTGTTTCAGACACTACGGATGATTTGTGGTTTTTAAATGAGACTGTGTCAGAGCAATTAG
GTGTTGAAGCTGCTAATTCTGAGCAAACAAGTGAAGTAGGGGAAACAAGTAACAAAGAGATGGGGGAGGTGGGAAAAGAT
GATGATCTTGAGGACTCCAAGTTCTTGAGCGAGGATACTGATATGGAACTTACCTCTGCGGATGAGTGGCAGTGTACGGA
ATGCAAGAAGTTTAATTCTCCAAGCAAGAGGTACTGTTTTCGTTGCTGGGCCTTGAGAAAGGATTGGTATTCGGATTGTT
CTAAATTAACTCATTCTCCGTCTACATCTAATATTACTGCCATACCTGAAAAGAAGGACAATGAAGAAATTGATGTTCCC
GATTATAGGAGAAACATTTCAGCTCCTGTTGTTAGGCCTAAAGATGGATATTTAAAGGAGGAAAAGCCCGGGTTCGACCC
TTGCAACTCAGTGGGAATTTTGGATTTGGCTCATAGTTCTAAAAGCCAGCAGATCATCTCAAGCGCGAGAGAACAACCAG
ATATTTTTTCTAAGCAGAAAGCTGAAACAGGAAGTATGGAAGATTTCCAGAATGTCTTGAAGCCGTGTAGCTTATGTGAA
AAAAGGCCTCGGGATGGGAACATTATTCATGGGAAGACGAGCCATCTGACAACATGTTTCCACTGTGCCAGAAGACTGAA
GAAGTCTGGGGCTTCGTGTCCTGTTTGTAACAAAGAGATTCAGTTGGTTATTAAAGTTTTTATAGCA
GGATGTCAGAACTTTCTGTGAAAGATCCAAGCCCTCTCTATGACGTGGCTAAGAAAGAATCTTCTATTCACACAGATGCT
GCTCAGACTCTCGCTCTCACACAGGATCACGCTATGGATTTCCCAAGTCAAGACCAACTGAAGCACGGCGCAACAGAGCA
CTCCGATCCCAGAAAAACAACTGAAGAAGAGGATACTCACACACTGCCTACCTCACCACGTAGATGCAGAGAGTCCAAAA
CAGATGAAGACTTGATAGAACATGTATCTCAAGATGAGACATCTAGGCTTGAACTTGATTTTGAGGAGTGGGACGTTGCT
GGCCTGCCTTGGTGGTTTCTAGGGAATTTGAGAAACGACGATATTCCTAAAAGTAATGGCTCAACTGATTTACAGACAAA
TCAGGATATAGGTACTGCAATTGTTTCAGACACTACGGATGATTTGTGGTTTTTAAATGAGACTGTGTCAGAGCAATTAG
GTGTTGAAGCTGCTAATTCTGAGCAAACAAGTGAAGTAGGGGAAACAAGTAACAAAGAGATGGGGGAGGTGGGAAAAGAT
GATGATCTTGAGGACTCCAAGTTCTTGAGCGAGGATACTGATATGGAACTTACCTCTGCGGATGAGTGGCAGTGTACGGA
ATGCAAGAAGTTTAATTCTCCAAGCAAGAGGTACTGTTTTCGTTGCTGGGCCTTGAGAAAGGATTGGTATTCGGATTGTT
CTAAATTAACTCATTCTCCGTCTACATCTAATATTACTGCCATACCTGAAAAGAAGGACAATGAAGAAATTGATGTTCCC
GATTATAGGAGAAACATTTCAGCTCCTGTTGTTAGGCCTAAAGATGGATATTTAAAGGAGGAAAAGCCCGGGTTCGACCC
TTGCAACTCAGTGGGAATTTTGGATTTGGCTCATAGTTCTAAAAGCCAGCAGATCATCTCAAGCGCGAGAGAACAACCAG
ATATTTTTTCTAAGCAGAAAGCTGAAACAGGAAGTATGGAAGATTTCCAGAATGTCTTGAAGCCGTGTAGCTTATGTGAA
AAAAGGCCTCGGGATGGGAACATTATTCATGGGAAGACGAGCCATCTGACAACATGTTTCCACTGTGCCAGAAGACTGAA
GAAGTCTGGGGCTTCGTGTCCTGTTTGTAACAAAGAGATTCAGTTGGTTATTAAAGTTTTTATAGCA
ORF - retro_mmus_3520 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 86.67 % |
| Parental protein coverage: | 82.62 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | GCQSF-SVKDPSPLYDMLRKNLVTSASINTDAAQTLALAQDHTMDFPSQDRLKHGATEYSNPRKRTEEED |
| GCQ.F.SVKDPSPLYD...K......SI.TDAAQTLAL.QDH.MDFPSQD.LKHGATE.S.PRK.TEEED | |
| Retrocopy | GCQNF<SVKDPSPLYDVAKKE----SSIHTDAAQTLALTQDHAMDFPSQDQLKHGATEHSDPRKTTEEED |
| Parental | THTLPTSRHKCRDSRADEDLIEHLSQDETSRLDLDFEEWDVAGLPWWFLGNLRNNCIPKSNGSTDLQTNQ |
| THTLPTS...CR.S..DEDLIEH.SQDETSRL.LDFEEWDVAGLPWWFLGNLRN..IPKSNGSTDLQTNQ | |
| Retrocopy | THTLPTSPRRCRESKTDEDLIEHVSQDETSRLELDFEEWDVAGLPWWFLGNLRNDDIPKSNGSTDLQTNQ |
| Parental | DIGTAIVSDTTDDLWFLNETVSEQLGVGIKVEAANSEQTSEVGKTSNKKTVEVGKDDDLEDSRSLSDDTD |
| DIGTAIVSDTTDDLWFLNETVSEQLG....VEAANSEQTSEVG.TSNK...EVGKDDDLEDS..LS.DTD | |
| Retrocopy | DIGTAIVSDTTDDLWFLNETVSEQLG----VEAANSEQTSEVGETSNKEMGEVGKDDDLEDSKFLSEDTD |
| Parental | VELTSEDEWQCTECKKFNSPSKRYCFRCWALRKDWYSDCSKLTHSLSTSNITAIPEKKDNEGIDVPDCRR |
| .ELTS.DEWQCTECKKFNSPSKRYCFRCWALRKDWYSDCSKLTHS.STSNITAIPEKKDNE.IDVPD.RR | |
| Retrocopy | MELTSADEWQCTECKKFNSPSKRYCFRCWALRKDWYSDCSKLTHSPSTSNITAIPEKKDNEEIDVPDYRR |
| Parental | TISAPVVRPKDGYLKEEKPRFDPCNSVGFLDLAHSSESQEIISSAREQTDIFSEQKAETESMEDFQNVLK |
| .ISAPVVRPKDGYLKEEKP.FDPCNSVG.LDLAHSS.SQ.IISSAREQ.DIFS.QKAET.SMEDFQNVLK | |
| Retrocopy | NISAPVVRPKDGYLKEEKPGFDPCNSVGILDLAHSSKSQQIISSAREQPDIFSKQKAETGSMEDFQNVLK |
| Parental | PCSLCEKRPRDGNIIHGKTSHLTTCFHCARRLKKSGASCPACKKEIQLVIKVFIA |
| PCSLCEKRPRDGNIIHGKTSHLTTCFHCARRLKKSGASCP.C.KEIQLVIKVFIA | |
| Retrocopy | PCSLCEKRPRDGNIIHGKTSHLTTCFHCARRLKKSGASCPVCNKEIQLVIKVFIA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 34 .46 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 44 .59 RPM |
| SRP007412_heart | 0 .00 RPM | 25 .12 RPM |
| SRP007412_kidney | 0 .02 RPM | 38 .61 RPM |
| SRP007412_liver | 0 .00 RPM | 25 .40 RPM |
| SRP007412_testis | 18 .39 RPM | 58 .64 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_3520 was not detected
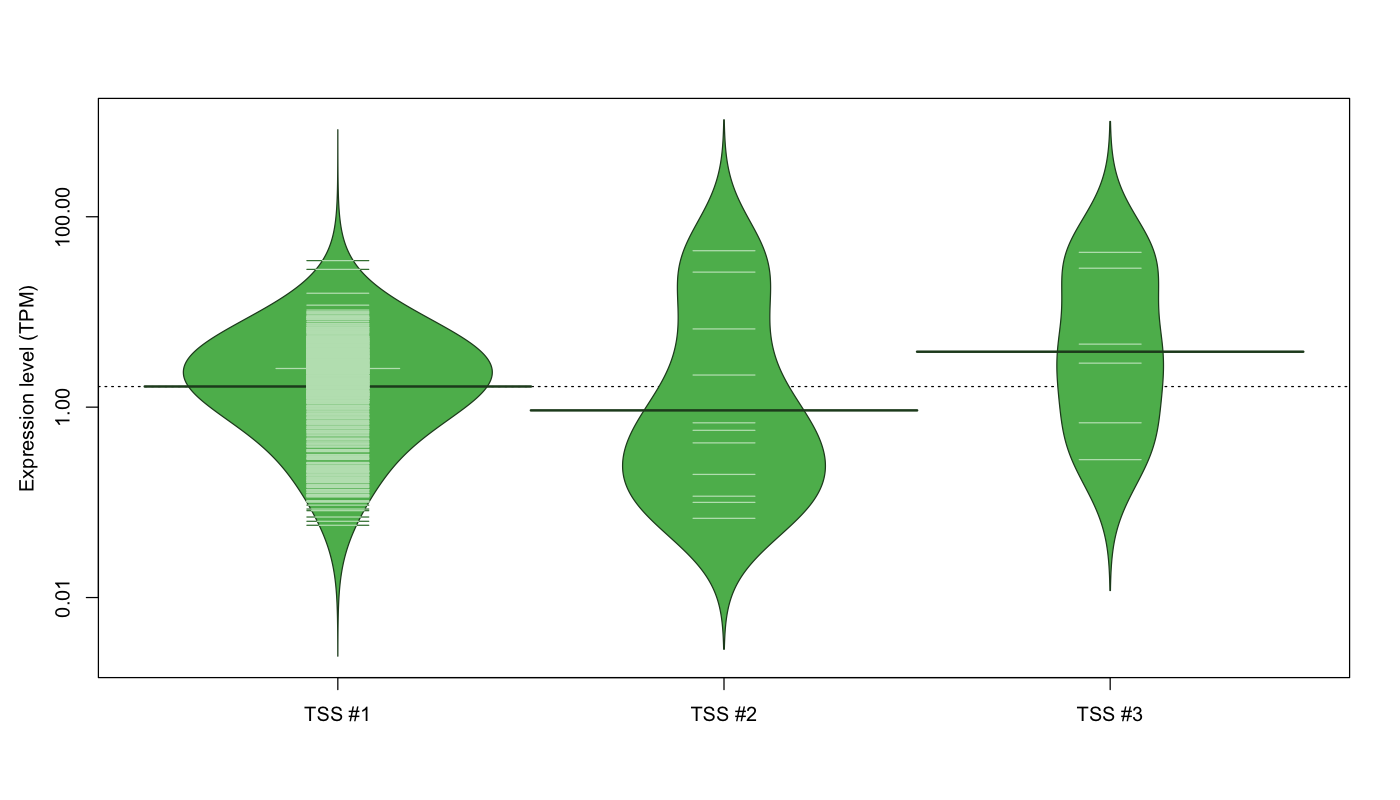
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_155101 | 370 libraries | 163 libraries | 465 libraries | 68 libraries | 6 libraries |
| TSS #2 | TSS_155102 | 1061 libraries | 7 libraries | 1 library | 1 library | 2 libraries |
| TSS #3 | TSS_155103 | 1066 libraries | 2 libraries | 2 libraries | 0 libraries | 2 libraries |
The graphical summary, for retro_mmus_3520 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3520 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3520 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 4 parental genes, and 4 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000001126 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000054387 | 1 retrocopy |
retro_mmus_3520 ,
|
| Otolemur garnettii | ENSOGAG00000011679 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000004184 | 1 retrocopy |