RetrogeneDB ID: | retro_mmus_460 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:80383106..80385149(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000090637 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Gle1 | ||
| Ensembl ID: | ENSMUSG00000019715 | ||
| Aliases: | None | ||
| Description: | GLE1 RNA export mediator (yeast) [Source:MGI Symbol;Acc:MGI:1921662] |

Retrocopy-Parental alignment summary:
>retro_mmus_460
CTCCGCAACTCCAGCAAAGGCCGTCTGCACTACAACCAGGATGTTTTGGAAGAATGCAGATCTCTCCCCAAACTATCTTC
TTACTCTGGATGGGTGGTAGATCACATTCTACCCAACACCTTACGCCACACTCAAGAGAATGGTCCTTCCTCGGATAACC
CATCATCCTCTGGATCTGCTTCAGGCCTGTACCAATCCTCACTACTCAAGTCTCCTGTCAGAAGTACCCTACAATCACTC
TCACCCTCAACATCGAATGGAACCCAGAACACACATGACTCCCGGTTCACAGAACCTATTGCACTTCAGTCCTCTCCAGC
GATCAAAGTGGAAGGTTGTATCCGAATGTTCGAATCGGCATCTGGAATAAGAGCAACAGAGGGCCTGACGAAGTGGCAGG
AGGAGCAAGAAAGGAAGGTGCAAGCCTTCTCAGAGATGGCATCCGAACAGCTAAAGAAGTTTGACGAGCTGAAGGAGTTG
AAGATGAATAAGGAGTTACGGGACTTAGAGGAAGTGATGGAGAAGAGTATCAGAGAAGCCCTGGGTCATCAGGAGAAGCT
GAAAGCTGAGCATCACCACAGAGCCAAGATTCTCAAGCTAATGCTTCAAGAGGCAGAGCAGCGGCAGCGTGAGAAGCAGG
CAGAGCAGGAGCAGCTTCGAAAGGAGGAAGGCCAGGCCCGTCTGCGCAGCCTGTACACCCTGCAAGAGGAGGTCCTGCAG
CTCAATCAGCAGCTGGATGCCTCCAGTCAACACAAAGACCTGCTGAATGTGGACCTGGCTGCCTTCCAGACCCAGGGCAA
CCAGCTGTGTAGCGTCATCTCCGGCATCATTTGCAGCACTTTGGAGAGTGGCATTCCCACGGCCGAGAACCAAGCTGAGG
CCGAGCTGGCCCTGCAGGAAATGCAGGACCTCCTCTTGAGTTTGGAGCAGGAGATCACCAGAGCCTCTGAAGTGAAGAAG
AAGAACGAGGAGGAAGAGGCCCAAGTCAAGCGGCAGGAGTCACAGATGCAGCAGGGGCCAGGAGTTCCCACCCAGAGCTC
TGCACCCAGCCCAAGCCCAACAGGTTCGCAGAATGAAGACCTGCAGGGGAAGGTACAAGATAGTACACTGCAGTGGTACC
AGCAACTTCAGGATGCTTCCGCCAAATGTGTGTTGGCCTTTGAGGACCTGACCAGCAGCAAGGACAGTCAGACGAAAAAG
ATAAGGACGGATTTGCGGAAGGCAGCCACCATTCCAGTGAGCCAGATCTCCGGCATCTCAGGTTCAAAGCTGAAGGAGGT
CTTTGATAAGCTCCACAGTCTGCTCTCGGGAAAGCCTGTCCAATCTGGAGGGCGTTCCGTGTCTGTCACTCTTAACCCTC
AGGGGCTGGACTTTGTCCAGTACAAACTGGCAGAGAAATTTGTGAAACAAGGGGAGAAGGAAGTGGCCTCTCACCACGAA
GCGGCATTCCCCATTGCGGCGGTGGCTTCTGGTATCTGGATGCTCCACCCCAAAGTGGGAGACCTCATTCTTGCTCATCT
CCACAAGAAGTGTCCATACTCCGTTCCTTTCTACCCAGCTTTCAAGGAAGGGATGGCTTTGGAGGATTATCAACGGATGC
TTGGCTATCAAGTTGTGGATTCCAAGGTTGAGCGGCAGGAAAGCTTTCTGAAACGCATGTCTGGGATGATCCGTTTGTAT
GCCGCCATCATCCAGCTCCAGTGGCCATATGCAAACCGGCAGGAGGTTCATCCTCATGGCTTAAATCATGGCTGGCGCTG
GTTGGCACAGATCTTAAACATGGAGCCTCTCTCAGGTGTAACAGCCACTCTCCTGTTTGACTTTTTGGAGGTGTGTGGGA
ACGCCCTCATGAAGCAGAACCAGGTCCAGTTCTGGAAGATGATACTTTTCATCCAAGAGGACTACTTGCCCAGAATTGAG
GCCATCACGAGCTCAGGACAGATGGGCTCTTTCATACGTCTCAAGCAGTTCTTGGAGAAGTGTCTGCAACGCAGGGAGAT
TCCTGTCCCCAGAGGCTTTCTGACGCCCTCCTTCTGGTGCTCT
CTCCGCAACTCCAGCAAAGGCCGTCTGCACTACAACCAGGATGTTTTGGAAGAATGCAGATCTCTCCCCAAACTATCTTC
TTACTCTGGATGGGTGGTAGATCACATTCTACCCAACACCTTACGCCACACTCAAGAGAATGGTCCTTCCTCGGATAACC
CATCATCCTCTGGATCTGCTTCAGGCCTGTACCAATCCTCACTACTCAAGTCTCCTGTCAGAAGTACCCTACAATCACTC
TCACCCTCAACATCGAATGGAACCCAGAACACACATGACTCCCGGTTCACAGAACCTATTGCACTTCAGTCCTCTCCAGC
GATCAAAGTGGAAGGTTGTATCCGAATGTTCGAATCGGCATCTGGAATAAGAGCAACAGAGGGCCTGACGAAGTGGCAGG
AGGAGCAAGAAAGGAAGGTGCAAGCCTTCTCAGAGATGGCATCCGAACAGCTAAAGAAGTTTGACGAGCTGAAGGAGTTG
AAGATGAATAAGGAGTTACGGGACTTAGAGGAAGTGATGGAGAAGAGTATCAGAGAAGCCCTGGGTCATCAGGAGAAGCT
GAAAGCTGAGCATCACCACAGAGCCAAGATTCTCAAGCTAATGCTTCAAGAGGCAGAGCAGCGGCAGCGTGAGAAGCAGG
CAGAGCAGGAGCAGCTTCGAAAGGAGGAAGGCCAGGCCCGTCTGCGCAGCCTGTACACCCTGCAAGAGGAGGTCCTGCAG
CTCAATCAGCAGCTGGATGCCTCCAGTCAACACAAAGACCTGCTGAATGTGGACCTGGCTGCCTTCCAGACCCAGGGCAA
CCAGCTGTGTAGCGTCATCTCCGGCATCATTTGCAGCACTTTGGAGAGTGGCATTCCCACGGCCGAGAACCAAGCTGAGG
CCGAGCTGGCCCTGCAGGAAATGCAGGACCTCCTCTTGAGTTTGGAGCAGGAGATCACCAGAGCCTCTGAAGTGAAGAAG
AAGAACGAGGAGGAAGAGGCCCAAGTCAAGCGGCAGGAGTCACAGATGCAGCAGGGGCCAGGAGTTCCCACCCAGAGCTC
TGCACCCAGCCCAAGCCCAACAGGTTCGCAGAATGAAGACCTGCAGGGGAAGGTACAAGATAGTACACTGCAGTGGTACC
AGCAACTTCAGGATGCTTCCGCCAAATGTGTGTTGGCCTTTGAGGACCTGACCAGCAGCAAGGACAGTCAGACGAAAAAG
ATAAGGACGGATTTGCGGAAGGCAGCCACCATTCCAGTGAGCCAGATCTCCGGCATCTCAGGTTCAAAGCTGAAGGAGGT
CTTTGATAAGCTCCACAGTCTGCTCTCGGGAAAGCCTGTCCAATCTGGAGGGCGTTCCGTGTCTGTCACTCTTAACCCTC
AGGGGCTGGACTTTGTCCAGTACAAACTGGCAGAGAAATTTGTGAAACAAGGGGAGAAGGAAGTGGCCTCTCACCACGAA
GCGGCATTCCCCATTGCGGCGGTGGCTTCTGGTATCTGGATGCTCCACCCCAAAGTGGGAGACCTCATTCTTGCTCATCT
CCACAAGAAGTGTCCATACTCCGTTCCTTTCTACCCAGCTTTCAAGGAAGGGATGGCTTTGGAGGATTATCAACGGATGC
TTGGCTATCAAGTTGTGGATTCCAAGGTTGAGCGGCAGGAAAGCTTTCTGAAACGCATGTCTGGGATGATCCGTTTGTAT
GCCGCCATCATCCAGCTCCAGTGGCCATATGCAAACCGGCAGGAGGTTCATCCTCATGGCTTAAATCATGGCTGGCGCTG
GTTGGCACAGATCTTAAACATGGAGCCTCTCTCAGGTGTAACAGCCACTCTCCTGTTTGACTTTTTGGAGGTGTGTGGGA
ACGCCCTCATGAAGCAGAACCAGGTCCAGTTCTGGAAGATGATACTTTTCATCCAAGAGGACTACTTGCCCAGAATTGAG
GCCATCACGAGCTCAGGACAGATGGGCTCTTTCATACGTCTCAAGCAGTTCTTGGAGAAGTGTCTGCAACGCAGGGAGAT
TCCTGTCCCCAGAGGCTTTCTGACGCCCTCCTTCTGGTGCTCT
ORF - retro_mmus_460 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 85.32 % |
| Parental protein coverage: | 97. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | LRYDREWLLRY-EDVLEECMSLPKLSSYSGWVVDHILPNTSGHTQESAPSSDNSPSSGSASGLYQSSLLK |
| LR......L.Y..DVLEEC.SLPKLSSYSGWVVDHILPNT..HTQE..PSSDN..SSGSASGLYQSSLLK | |
| Retrocopy | LRNSSKGRLHYNQDVLEECRSLPKLSSYSGWVVDHILPNTLRHTQENGPSSDNPSSSGSASGLYQSSLLK |
| Parental | SPVRSSPQSPSPSSPNGTQSTHESPFTEPIAPQSSRAIKVEGCIRMYELAHRMRGTEGLRQWQEEQERKV |
| SPVRS..QS.SPS..NGTQ.TH.S.FTEPIA.QSS.AIKVEGCIRM.E.A...R.TEGL..WQEEQERKV | |
| Retrocopy | SPVRSTLQSLSPSTSNGTQNTHDSRFTEPIALQSSPAIKVEGCIRMFESASGIRATEGLTKWQEEQERKV |
| Parental | RALSEMASEQLKRFDELKELKLHKEFQDLQEVMEKSTREALGHQEKLKEEHRHRAKILNLKLREAEQ-QR |
| .A.SEMASEQLK.FDELKELK..KE..DL.EVMEKS.REALGHQEKLK.EH.HRAKIL.L.L.EAEQ.QR | |
| Retrocopy | QAFSEMASEQLKKFDELKELKMNKELRDLEEVMEKSIREALGHQEKLKAEHHHRAKILKLMLQEAEQRQR |
| Parental | VKQAEQEQLRKEEGQVRLRSLYSLQEEVLQLNQQLDASSQHKELLSVDLAAFQTRGNQLCGLISSIIRTT |
| .KQAEQEQLRKEEGQ.RLRSLY.LQEEVLQLNQQLDASSQHK.LL.VDLAAFQT.GNQLC..IS.II..T | |
| Retrocopy | EKQAEQEQLRKEEGQARLRSLYTLQEEVLQLNQQLDASSQHKDLLNVDLAAFQTQGNQLCSVISGIICST |
| Parental | LESGYPTAENQAEAERALQEMRDLLSDLEQEITRASQVKKKHEEE-AKVKRQESQVQQGPAPPTQTSAPS |
| LESG.PTAENQAEAE.ALQEM.DLL..LEQEITRAS.VKKK.EEE.A.VKRQESQ.QQGP..PTQ.SAPS | |
| Retrocopy | LESGIPTAENQAEAELALQEMQDLLLSLEQEITRASEVKKKNEEEEAQVKRQESQMQQGPGVPTQSSAPS |
| Parental | PSPVGAQNEDLQVKVQDSTMQWYQQLQDASAKCVLAFEDLTSSKDSQTKKIKMDLQKAATIPVSQISTIA |
| PSP.G.QNEDLQ.KVQDST.QWYQQLQDASAKCVLAFEDLTSSKDSQTKKI..DL.KAATIPVSQIS.I. | |
| Retrocopy | PSPTGSQNEDLQGKVQDSTLQWYQQLQDASAKCVLAFEDLTSSKDSQTKKIRTDLRKAATIPVSQISGIS |
| Parental | GSKLKEIFDKIHSLLSGKPVQSGGRSVSVTLNPQGLDFVQYKLAEKFVKQGEEEVASHHEAAFPIAVVAS |
| GSKLKE.FDK.HSLLSGKPVQSGGRSVSVTLNPQGLDFVQYKLAEKFVKQGE.EVASHHEAAFPIA.VAS | |
| Retrocopy | GSKLKEVFDKLHSLLSGKPVQSGGRSVSVTLNPQGLDFVQYKLAEKFVKQGEKEVASHHEAAFPIAAVAS |
| Parental | GIWMLHPKVGDLILAHLHKKCPYSVPFYPAFKEGMALEDYQRMLGYQVTDSKVEQQDNFLKRMSGMIRLY |
| GIWMLHPKVGDLILAHLHKKCPYSVPFYPAFKEGMALEDYQRMLGYQV.DSKVE.Q..FLKRMSGMIRLY | |
| Retrocopy | GIWMLHPKVGDLILAHLHKKCPYSVPFYPAFKEGMALEDYQRMLGYQVVDSKVERQESFLKRMSGMIRLY |
| Parental | AAIIQLQWPYGNRQEAHPHGLNHGWRWLAQVLNMEPLSDVTATLLFDFLEVCGNALMKQYQVQFWKMILL |
| AAIIQLQWPY.NRQE.HPHGLNHGWRWLAQ.LNMEPLS.VTATLLFDFLEVCGNALMKQ.QVQFWKMIL. | |
| Retrocopy | AAIIQLQWPYANRQEVHPHGLNHGWRWLAQILNMEPLSGVTATLLFDFLEVCGNALMKQNQVQFWKMILF |
| Parental | IKEDYFPRIEAITSSGQMGSFIRLKQFLEKCLQRREIPVPRGFLTTSFWRS |
| I.EDY.PRIEAITSSGQMGSFIRLKQFLEKCLQRREIPVPRGFLT.SFW.S | |
| Retrocopy | IQEDYLPRIEAITSSGQMGSFIRLKQFLEKCLQRREIPVPRGFLTPSFWCS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 47 .29 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 67 .00 RPM |
| SRP007412_heart | 0 .06 RPM | 31 .88 RPM |
| SRP007412_kidney | 0 .10 RPM | 39 .17 RPM |
| SRP007412_liver | 0 .00 RPM | 33 .51 RPM |
| SRP007412_testis | 0 .09 RPM | 41 .35 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_460 was not detected
No EST(s) were mapped for retro_mmus_460 retrocopy.
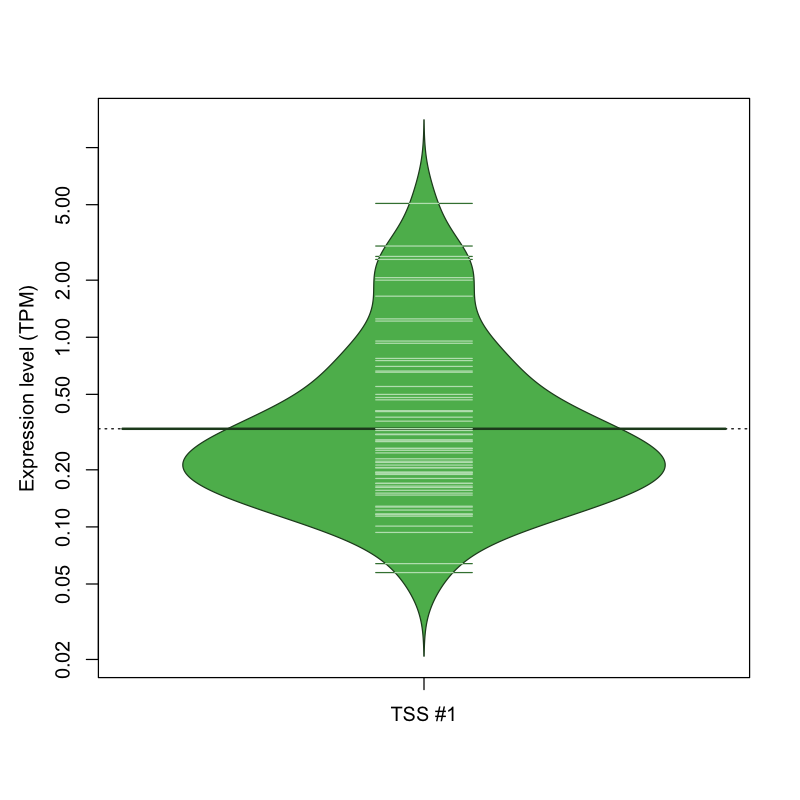
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_76680 | 1005 libraries | 58 libraries | 8 libraries | 1 library | 0 libraries |
The graphical summary, for retro_mmus_460 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_460 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_460 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 4 parental genes, and 5 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Microcebus murinus | ENSMICG00000017640 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000007379 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000019715 | 2 retrocopies |
retro_mmus_3505, retro_mmus_460 ,
|
| Rattus norvegicus | ENSRNOG00000015237 | 1 retrocopy |