RetrogeneDB ID: | retro_mmus_72 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 10:116112482..116113619(-) | ||
| Located in intron of: | ENSMUSG00000020151 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000074734 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Taf7l | ||
| Ensembl ID: | ENSMUSG00000009596 | ||
| Aliases: | Taf7l, 4933438I11Rik, 50kDa, Taf2q | ||
| Description: | TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor [Source:MGI Symbol;Acc:MGI:1921719] |

Retrocopy-Parental alignment summary:
>retro_mmus_72
ATGAGCAAAAGCCGAGATGAACCGCCTCACGAACTCGAGAACCAGTTCATACTGCGGCTGCCCCCAGAACAGGCTTCGGC
GGTCAGGAAGATCATCCGTTCTGGAAATGCCGCCATGAGAGAGAAACTAAAAATCGACTTGTCTCCCGATTCACGCCGTG
CCGTCGTTCAGGTAGATGGCGTCTCACTGTCTGCCAAACTTGTCGATTTGCCTTGTGTTATTGGAAGCCTGAAAACGCAC
GATGGGAAAACCTTTTATAAAACAGCCGATGTGTCCCAGATGCTATTGTGCAGCGCCGATGATGGTGATCCCCACACGTC
CCCAGAAGAGCCAGCTACCTCTGCTGGTGCTACTGTAGTGGGAAACAAAAAGGAAGCAGAGGGAAAATATATCTGGAAGC
ATGGCATTACTCCACCACTTAAGAACGTCAGAAAGAAACGGTTTCGCAAACCAACAAAAAAGCCCGCCGATATGAAGCAA
GGCGAAGAAAGCTGTAGGGCCTACATTGATTCCAAAGATGTGGAAAAGGAAGTGAAGAGGCTGCTGCGCTCAGATGCTGA
AGCCATCAGCAGCCGATGGGAAGTCGTTGTTGATGATGAAACCAAGGCAGTACCAAGTCAAACCTGCACTTCACACGTTC
CGTTGCCCCCTGAAGCAGGTGATCATACCTCATCAGCATGTGCTATGCCTCAGAGCACATTCAGTGATTCCAGCAGTGGC
AAAGATGATCCAGAGAATAAAGATGAGGAGGAGGAAGAAGAAAAAGAGGAGAAGGAAGAAGAGGATGAGGATGAGGAGGA
TGAAGAGGAGGAAGAGGAAGAAGAGGAAGATGATTCTGAAGAAGACCTGGAAAGAGAGCTACAAGCCAAGTTTATTGAGT
TTAGCTTGTGTGAAGCTAATGAAAGTAGCAGTTCAGTAATTCTTGGAATTCAGAAGCTGATTCATTCCAAGGAGAGAAAA
CTCCAAGAGATCCAGGCCAAAGCACAGAGGCAGAAAGATCTCCTCAGAAAATTGGAAAACCTGACCCTCAAGAGTCATTT
CCAGTCTGTGCTAGATCAGCTCAAGTTACAGGAGAAGCAGAAGTATGAACAGATCCTTTTTCTACAGGAACAACTGAAGT
GTTTTTTGAAGAAGTAA
ATGAGCAAAAGCCGAGATGAACCGCCTCACGAACTCGAGAACCAGTTCATACTGCGGCTGCCCCCAGAACAGGCTTCGGC
GGTCAGGAAGATCATCCGTTCTGGAAATGCCGCCATGAGAGAGAAACTAAAAATCGACTTGTCTCCCGATTCACGCCGTG
CCGTCGTTCAGGTAGATGGCGTCTCACTGTCTGCCAAACTTGTCGATTTGCCTTGTGTTATTGGAAGCCTGAAAACGCAC
GATGGGAAAACCTTTTATAAAACAGCCGATGTGTCCCAGATGCTATTGTGCAGCGCCGATGATGGTGATCCCCACACGTC
CCCAGAAGAGCCAGCTACCTCTGCTGGTGCTACTGTAGTGGGAAACAAAAAGGAAGCAGAGGGAAAATATATCTGGAAGC
ATGGCATTACTCCACCACTTAAGAACGTCAGAAAGAAACGGTTTCGCAAACCAACAAAAAAGCCCGCCGATATGAAGCAA
GGCGAAGAAAGCTGTAGGGCCTACATTGATTCCAAAGATGTGGAAAAGGAAGTGAAGAGGCTGCTGCGCTCAGATGCTGA
AGCCATCAGCAGCCGATGGGAAGTCGTTGTTGATGATGAAACCAAGGCAGTACCAAGTCAAACCTGCACTTCACACGTTC
CGTTGCCCCCTGAAGCAGGTGATCATACCTCATCAGCATGTGCTATGCCTCAGAGCACATTCAGTGATTCCAGCAGTGGC
AAAGATGATCCAGAGAATAAAGATGAGGAGGAGGAAGAAGAAAAAGAGGAGAAGGAAGAAGAGGATGAGGATGAGGAGGA
TGAAGAGGAGGAAGAGGAAGAAGAGGAAGATGATTCTGAAGAAGACCTGGAAAGAGAGCTACAAGCCAAGTTTATTGAGT
TTAGCTTGTGTGAAGCTAATGAAAGTAGCAGTTCAGTAATTCTTGGAATTCAGAAGCTGATTCATTCCAAGGAGAGAAAA
CTCCAAGAGATCCAGGCCAAAGCACAGAGGCAGAAAGATCTCCTCAGAAAATTGGAAAACCTGACCCTCAAGAGTCATTT
CCAGTCTGTGCTAGATCAGCTCAAGTTACAGGAGAAGCAGAAGTATGAACAGATCCTTTTTCTACAGGAACAACTGAAGT
GTTTTTTGAAGAAGTAA
ORF - retro_mmus_72 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 60.22 % |
| Parental protein coverage: | 78.13 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | HDVEEQFILRLPPEQAYAVRKIIHSRNAAWKDKLKIDFSPDGHHAVVQVDNVSLPAKLVNLPCVIGSLKT |
| H..E.QFILRLPPEQA.AVRKII.S.NAA...KLKID.SPD...AVVQVD.VSL.AKLV.LPCVIGSLKT | |
| Retrocopy | HELENQFILRLPPEQASAVRKIIRSGNAAMREKLKIDLSPDSRRAVVQVDGVSLSAKLVDLPCVIGSLKT |
| Parental | IDRKTFYKTADVSQMLVCS-PEGEPHSPPEEPVVSTGPTVIGISEGKAERKKYNWKHGITPPLKNVRKKR |
| .D.KTFYKTADVSQML.CS...G.PH..PEEP..S.G.TV.G....K....KY.WKHGITPPLKNVRKKR | |
| Retrocopy | HDGKTFYKTADVSQMLLCSADDGDPHTSPEEPATSAGATVVG--NKKEAEGKYIWKHGITPPLKNVRKKR |
| Parental | FRKTTKKLPDVKQVDEINFSEYTQSPSVEKEVKRLLYSDAEAVSVRWEVVDDDDAKEIESQGSMPTTPGI |
| FRK.TKK..D.KQ..E.....Y..S..VEKEVKRLL.SDAEA.S.RWEVV.DD..K...SQ......P.. | |
| Retrocopy | FRKPTKKPADMKQGEE-SCRAYIDSKDVEKEVKRLLRSDAEAISSRWEVVVDDETKAVPSQTCTSHVPLP |
| Parental | SQMGGASLSDYDVFREMMGDSGSNSNDVEEKSNEGDDDDDE---DEDDEDYGNEKEEEETDNSEEELEKE |
| ...G....S..........DS.S...D.E.K..E......E......DE....E.EEEE.D.SEE.LE.E | |
| Retrocopy | PEAGDHTSSACAMPQSTFSDSSSGKDDPENKDEEEEEEKEEKEEEDEDEEDEEEEEEEEEDDSEEDLERE |
| Parental | LQAKFNEFSLHEADQDYSSITMAIQKLIFIKEKRLQMIYKKAQRQKELLRKVENLTLKRHFQNVLGKLNI |
| LQAKF.EFSL.EA....SS....IQKLI..KE..LQ.I..KAQRQK.LLRK.ENLTLK.HFQ.VL..L.. | |
| Retrocopy | LQAKFIEFSLCEANESSSSVILGIQKLIHSKERKLQEIQAKAQRQKDLLRKLENLTLKSHFQSVLDQLKL |
| Parental | MEKEKCEQIYHLQEQLKCFLKE |
| .EK.K.EQI..LQEQLKCFLK. | |
| Retrocopy | QEKQKYEQILFLQEQLKCFLKK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 0 .28 RPM |
| SRP007412_cerebellum | 0 .13 RPM | 0 .04 RPM |
| SRP007412_heart | 0 .00 RPM | 0 .00 RPM |
| SRP007412_kidney | 0 .00 RPM | 0 .02 RPM |
| SRP007412_liver | 0 .03 RPM | 0 .00 RPM |
| SRP007412_testis | 110 .00 RPM | 27 .08 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_72 was not detected
No EST(s) were mapped for retro_mmus_72 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_521 | 1066 libraries | 1 library | 2 libraries | 1 library | 2 libraries |
| TSS #2 | TSS_522 | 1062 libraries | 3 libraries | 1 library | 2 libraries | 4 libraries |
| TSS #3 | TSS_523 | 1066 libraries | 2 libraries | 1 library | 2 libraries | 1 library |
| TSS #4 | TSS_524 | 1063 libraries | 5 libraries | 2 libraries | 1 library | 1 library |
| TSS #5 | TSS_525 | 1064 libraries | 4 libraries | 1 library | 2 libraries | 1 library |
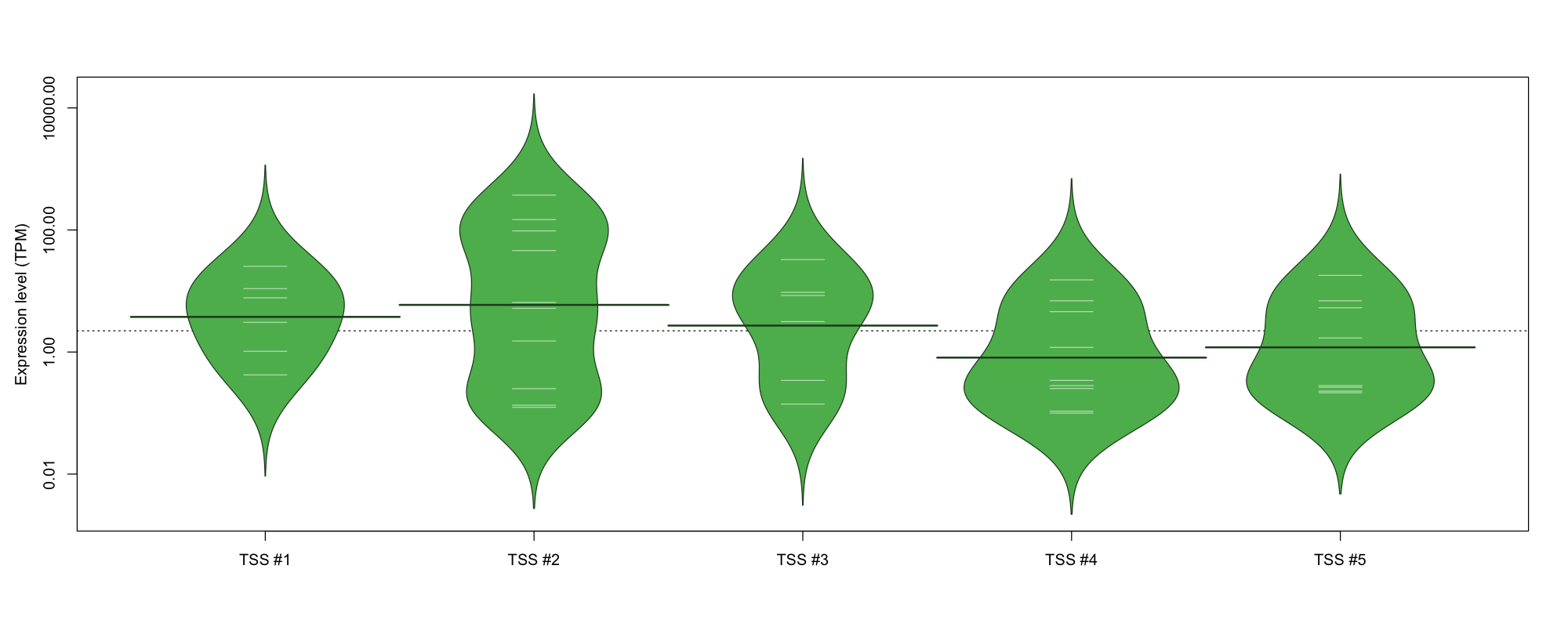
The graphical summary, for retro_mmus_72 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_72 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_72 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_134 |
Parental genes homology:
Parental genes homology involve 6 parental genes, and 6 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000009247 | 1 retrocopy | |
| Bos taurus | ENSBTAG00000001979 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000006941 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000009596 | 1 retrocopy |
retro_mmus_72 ,
|
| Oryctolagus cuniculus | ENSOCUG00000004497 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000026315 | 1 retrocopy |