RetrogeneDB ID: | retro_mmus_106 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 17:33065099..33067826(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000023350 | |
| Aliases: | 4921501E09Rik, PHF8 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Phf8 | ||
| Ensembl ID: | ENSMUSG00000041229 | ||
| Aliases: | Phf8, 9830141C09Rik, mKIAA1111 | ||
| Description: | PHD finger protein 8 [Source:MGI Symbol;Acc:MGI:2444341] |

Retrocopy-Parental alignment summary:
>retro_mmus_106
ATGGCCTTGGTGCCTGTGTATTGTCTTTGTCGACAGCCTTATAATGTGAACCACTTCATGATTGAGTGCGGCCTGTGCCA
GGACTGGTTTCATGGTAGCTGTGTTGGGATTGAAGAAGAAAACGCTGTTGACATTGACATCTACCACTGCCCAGACTGTG
AGGCTGTATTTGGGCCTTCCATTATGAAAAACTGGCATGGATCGCCTAGAGGGCATGATGCTCTCAAGAGGAAGCCATTG
ATGACTGGGAGCCCTACCTCTATTCACCAAATCCAGGGTAGGATTTTTGAAAGCTCTGACAAAGTGATTTTGAAGCCCAC
AGGAAGTCATTTAACAGTAGAATTTCTGCTAGAAAGTAGCTTCAGCATACCTATCCTTGTCTTGAAGAAGGATGGGTTGG
GGATGACACTTCCACCGACATCATTCACTGTCAGAGATGTTGAGCAATATGTTGGTTCTGACAGAGAGGTTGATGTGATT
GATGTAGCCTGTCAGGCTAGCAGCAAGATGATGTTGGGAGATTTTGTGAAATATTATTACAGCAGGAAGAGGGAAAAGAC
CTTTAATGTTATTAATTTGGAATTCTCTAATACTGGGCTTTCTAATATAGTAGAGATGCCCAAGATAGTTCGTAAGCTTT
CCTGGGTTGAAAACTTATGGCCAGAAGATTGCGGCTTTGAGAGACCCAATGTACAGAAGTATTGCCTTATGAGTATGCAG
AATAGCTATACAGATTTTCACATTGAATTTGGTGGCACTTCAGCCTGGTACCATGTAGTTAAGGGTGAGAAGATCTTCTA
TCTGATCCGCCCAACAGATGCTAACCTGACTCTTTTTGAGTGTTGGAGCAACTCTTCTAACCAAAAGGAAATATTCTTTG
GTGACCAGGTAGACAGATGTTACAAGTGTTCTGTGAAGCAAGGACAGACACTTTTCATTCCTACAGGATGGATCTATGCT
GTGCTGACCCCAGTGGATTGCCTAGCATTTGGGGGAAACTTCTTACACAGTTTAAACATTGAAATGCAGCTCAAAGTCTA
TGAGATTGAGAAGCATCTGTGTACAACAGACCTCTTCAAGTTTCCAAGCTTTGAGACCATCTGTTGGCATGTGGGGAAAC
ACATCCTGGACATCTTTAAAGGTTTACGGGAGAATGGGAGATGTCCTGCATCCTTCTTGGTCCATGGTGGTAAAGCTCTA
AACTTGGCCTTTCAAGCCTGGACAAGGAAAGAAGTTCTGCCTAACCGTGATAATGAGATCCCAAAGACAGTGCAGACCAT
ACAGCTCATTGAAGATCTGGCTTGGGAAATTCATCTGGTTGAAGATAATTTCCAACAGAACATCCCAAAGACAAGTTCTA
CCTTTAAGTTCCAGCAGCTCCATACCAATTCCTCTTCCTTAACTAGGCAAGCTCATCCCACTTCAGTATCCAAGTCTAAA
TTATCACTGCCCTCCAAAAATGGTTCAGAAAAGAAAAGCCCAAAGTATAAAGACACCTTTAAGAAGGAAGAACAAATGAG
CAAGGAGAGTTCAGCTCTGGGGCCTAATTATCAACATAGCTGTAATTTCATGGATCTAAATAATGATCAACAATGGAAGG
TAGGCTGTCTCCAAAACGTAGAGTTTAACATCACTAGTACTTGTTTGAATGATTCAGATGATAACTCAAAACATGACAAA
GCCTTCAATGGGAATGAGAGCCTGGTGCCTCTGTTGATGGCTAATGGCAATACGAAGAGGGAAAAGAGCTTGTCCGAATC
TTGGCAAGCCAAAATCACAAAGAAGGAAGATACATCAAGGTTGATGAAAGAACAAGCGATGGTAGACACATTTGACTTGG
ATTTAAATGATGAGCTACAGATTGAAAAGGGATTAGGTAAAGAGAAAGTGAGTCTGGTAGTAAGACCAAAAATTCCTCCA
AGTTTGCCCCATGTGAAGCTTTGCTCTGACCAAAACCAAAGTCATGATCCAGGGGAAACTGAGATCACCATTGTAGAGAA
CTATAAAATAGATAAGGAAATGGTGGAAGAGGTTGAAGGCAAGTTTAAGAATGGAAGTGAATCTGGTGGAACTATTGATC
CACTAAAGGCCAAGAAACAGGTGGGTGGACCTGGCTGTGTTATCCTCACTGATGACCCAAACTCTTCCAGCAATAAGAAG
GACATTCAGGACACTGTGTGCATGTCTGATTTGCAGTCTTCTCCTCCTTCACAAGCTCCCTCCAGCCTTCAGGCATTTTG
GAGTGGGGGACAAGACCAAGGCAACAAAAGCCCTAGCAGTGTCTTGGGCACAGTGTCTGACAGTCCTGTTTCTCGGCGTA
CCCCAAGGAAATGGCCCATCAAGCGGCTAGCATACTGGAGAAATGAAAGTGAGGCGGGAAATGCTTGTGTAGATGAACAG
GGCAGCTTAGGAGCATGCTTCAAGGAAGCTAAGTATATCTTCTTATCCCTGGAATCTGATGACGACGACGATGATGATGA
TGATGACGACGATGACAACAACCCTGCTTTGAGAGTTCTACCAAAGAAAAGGAAGATATCAGATAATGCCCCAAGGATTC
CTAGGGCAGATGAGATCCCAACACTACCAAAGCAGGACCATTCTGTGCCTGAGGGTAACAGAGTTGGCTCCATTGAGATA
GACTGGACTACAGCAGCTACAAAGCTGGCACAGGAGGTGAACAACATGAAGCTCACTCTCACTAGTCCAGGAAAAAGCAG
CCAATAA
ATGGCCTTGGTGCCTGTGTATTGTCTTTGTCGACAGCCTTATAATGTGAACCACTTCATGATTGAGTGCGGCCTGTGCCA
GGACTGGTTTCATGGTAGCTGTGTTGGGATTGAAGAAGAAAACGCTGTTGACATTGACATCTACCACTGCCCAGACTGTG
AGGCTGTATTTGGGCCTTCCATTATGAAAAACTGGCATGGATCGCCTAGAGGGCATGATGCTCTCAAGAGGAAGCCATTG
ATGACTGGGAGCCCTACCTCTATTCACCAAATCCAGGGTAGGATTTTTGAAAGCTCTGACAAAGTGATTTTGAAGCCCAC
AGGAAGTCATTTAACAGTAGAATTTCTGCTAGAAAGTAGCTTCAGCATACCTATCCTTGTCTTGAAGAAGGATGGGTTGG
GGATGACACTTCCACCGACATCATTCACTGTCAGAGATGTTGAGCAATATGTTGGTTCTGACAGAGAGGTTGATGTGATT
GATGTAGCCTGTCAGGCTAGCAGCAAGATGATGTTGGGAGATTTTGTGAAATATTATTACAGCAGGAAGAGGGAAAAGAC
CTTTAATGTTATTAATTTGGAATTCTCTAATACTGGGCTTTCTAATATAGTAGAGATGCCCAAGATAGTTCGTAAGCTTT
CCTGGGTTGAAAACTTATGGCCAGAAGATTGCGGCTTTGAGAGACCCAATGTACAGAAGTATTGCCTTATGAGTATGCAG
AATAGCTATACAGATTTTCACATTGAATTTGGTGGCACTTCAGCCTGGTACCATGTAGTTAAGGGTGAGAAGATCTTCTA
TCTGATCCGCCCAACAGATGCTAACCTGACTCTTTTTGAGTGTTGGAGCAACTCTTCTAACCAAAAGGAAATATTCTTTG
GTGACCAGGTAGACAGATGTTACAAGTGTTCTGTGAAGCAAGGACAGACACTTTTCATTCCTACAGGATGGATCTATGCT
GTGCTGACCCCAGTGGATTGCCTAGCATTTGGGGGAAACTTCTTACACAGTTTAAACATTGAAATGCAGCTCAAAGTCTA
TGAGATTGAGAAGCATCTGTGTACAACAGACCTCTTCAAGTTTCCAAGCTTTGAGACCATCTGTTGGCATGTGGGGAAAC
ACATCCTGGACATCTTTAAAGGTTTACGGGAGAATGGGAGATGTCCTGCATCCTTCTTGGTCCATGGTGGTAAAGCTCTA
AACTTGGCCTTTCAAGCCTGGACAAGGAAAGAAGTTCTGCCTAACCGTGATAATGAGATCCCAAAGACAGTGCAGACCAT
ACAGCTCATTGAAGATCTGGCTTGGGAAATTCATCTGGTTGAAGATAATTTCCAACAGAACATCCCAAAGACAAGTTCTA
CCTTTAAGTTCCAGCAGCTCCATACCAATTCCTCTTCCTTAACTAGGCAAGCTCATCCCACTTCAGTATCCAAGTCTAAA
TTATCACTGCCCTCCAAAAATGGTTCAGAAAAGAAAAGCCCAAAGTATAAAGACACCTTTAAGAAGGAAGAACAAATGAG
CAAGGAGAGTTCAGCTCTGGGGCCTAATTATCAACATAGCTGTAATTTCATGGATCTAAATAATGATCAACAATGGAAGG
TAGGCTGTCTCCAAAACGTAGAGTTTAACATCACTAGTACTTGTTTGAATGATTCAGATGATAACTCAAAACATGACAAA
GCCTTCAATGGGAATGAGAGCCTGGTGCCTCTGTTGATGGCTAATGGCAATACGAAGAGGGAAAAGAGCTTGTCCGAATC
TTGGCAAGCCAAAATCACAAAGAAGGAAGATACATCAAGGTTGATGAAAGAACAAGCGATGGTAGACACATTTGACTTGG
ATTTAAATGATGAGCTACAGATTGAAAAGGGATTAGGTAAAGAGAAAGTGAGTCTGGTAGTAAGACCAAAAATTCCTCCA
AGTTTGCCCCATGTGAAGCTTTGCTCTGACCAAAACCAAAGTCATGATCCAGGGGAAACTGAGATCACCATTGTAGAGAA
CTATAAAATAGATAAGGAAATGGTGGAAGAGGTTGAAGGCAAGTTTAAGAATGGAAGTGAATCTGGTGGAACTATTGATC
CACTAAAGGCCAAGAAACAGGTGGGTGGACCTGGCTGTGTTATCCTCACTGATGACCCAAACTCTTCCAGCAATAAGAAG
GACATTCAGGACACTGTGTGCATGTCTGATTTGCAGTCTTCTCCTCCTTCACAAGCTCCCTCCAGCCTTCAGGCATTTTG
GAGTGGGGGACAAGACCAAGGCAACAAAAGCCCTAGCAGTGTCTTGGGCACAGTGTCTGACAGTCCTGTTTCTCGGCGTA
CCCCAAGGAAATGGCCCATCAAGCGGCTAGCATACTGGAGAAATGAAAGTGAGGCGGGAAATGCTTGTGTAGATGAACAG
GGCAGCTTAGGAGCATGCTTCAAGGAAGCTAAGTATATCTTCTTATCCCTGGAATCTGATGACGACGACGATGATGATGA
TGATGACGACGATGACAACAACCCTGCTTTGAGAGTTCTACCAAAGAAAAGGAAGATATCAGATAATGCCCCAAGGATTC
CTAGGGCAGATGAGATCCCAACACTACCAAAGCAGGACCATTCTGTGCCTGAGGGTAACAGAGTTGGCTCCATTGAGATA
GACTGGACTACAGCAGCTACAAAGCTGGCACAGGAGGTGAACAACATGAAGCTCACTCTCACTAGTCCAGGAAAAAGCAG
CCAATAA
ORF - retro_mmus_106 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 68.61 % |
| Parental protein coverage: | 80.55 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MASVPVYCLCRLPYDVTRFMIECDMCQDWFHGSCVGVEEEKAADIDLYHCPNCEVLHGPSIMKKRRGSSK |
| MA.VPVYCLCR.PY.V..FMIEC..CQDWFHGSCVG.EEE.A.DID.YHCP.CE...GPSIMK...GS.. | |
| Retrocopy | MALVPVYCLCRQPYNVNHFMIECGLCQDWFHGSCVGIEEENAVDIDIYHCPDCEAVFGPSIMKNWHGSPR |
| Parental | GHDNHKGKPLKTGSSMFIRELRGRTFDSSDEVILKPTGSQLTVEFLEENSFSVPILVLKKDGLGMTLPSP |
| GHD..K.KPL.TGS...I....GR.F.SSD.VILKPTGS.LTVEFL.E.SFS.PILVLKKDGLGMTLP.. | |
| Retrocopy | GHDALKRKPLMTGSPTSIHQIQGRIFESSDKVILKPTGSHLTVEFLLESSFSIPILVLKKDGLGMTLPPT |
| Parental | SFTVRDVEHYVGSDKEIDVIDVARQADCKMKLGDFVKYYYSGKREKVLNVISLEFSDTRLSNLVETPRIV |
| SFTVRDVE.YVGSD.E.DVIDVA.QA..KM.LGDFVKYYYS.KREK..NVI.LEFS.T.LSN.VE.P.IV | |
| Retrocopy | SFTVRDVEQYVGSDREVDVIDVACQASSKMMLGDFVKYYYSRKREKTFNVINLEFSNTGLSNIVEMPKIV |
| Parental | RKLSWVENLWPEECVFERPNVQKYCLMSVRDSYTDFHIDFGGTSVWYHVLKGEKIFYLIRPTNANLTLFE |
| RKLSWVENLWPE.C.FERPNVQKYCLMS...SYTDFHI.FGGTS.WYHV.KGEKIFYLIRPT.ANLTLFE | |
| Retrocopy | RKLSWVENLWPEDCGFERPNVQKYCLMSMQNSYTDFHIEFGGTSAWYHVVKGEKIFYLIRPTDANLTLFE |
| Parental | CWSSSSNQNEMFFGDQVEKCYKCSVKQGQTLFIPTGWIHAVLTPVDCLAFGGNFLHSLNIEMQLKAYEIE |
| CWS.SSNQ.E.FFGDQV..CYKCSVKQGQTLFIPTGWI.AVLTPVDCLAFGGNFLHSLNIEMQLK.YEIE | |
| Retrocopy | CWSNSSNQKEIFFGDQVDRCYKCSVKQGQTLFIPTGWIYAVLTPVDCLAFGGNFLHSLNIEMQLKVYEIE |
| Parental | KRLSTADLFKFPNFETICWYVGKHILDIFRGLRENRRHPASYLVHGGKALNLAFRAWTKKEALPDHEDEI |
| K.L.T.DLFKFP.FETICW.VGKHILDIF.GLREN.R.PAS.LVHGGKALNLAF.AWT.KE.LP....EI | |
| Retrocopy | KHLCTTDLFKFPSFETICWHVGKHILDIFKGLRENGRCPASFLVHGGKALNLAFQAWTRKEVLPNRDNEI |
| Parental | PETVRTVQLIKDLAREIRLVEDIFQQNVGKTSNIFGLQRIFPAGSIPLTKPAHSTSVSMSKLSLPSKNGS |
| P.TV.T.QLI.DLA.EI.LVED.FQQN..KTS..F..Q......S..LT..AH.TSVS.SKLSLPSKNGS | |
| Retrocopy | PKTVQTIQLIEDLAWEIHLVEDNFQQNIPKTSSTFKFQQLH-TNSSSLTRQAHPTSVSKSKLSLPSKNGS |
| Parental | KKKGLKPKDIFKKAERKGKQSSALGPAGQLSYNLMDPYSHQALKTGPSQKAKFNMSGTSLNDSDDDSA-D |
| .KK..K.KD.FKK.E...K.SSALGP..Q.S.N.MD....Q..K.G..Q...FN...T.LNDSDD.S..D | |
| Retrocopy | EKKSPKYKDTFKKEEQMSKESSALGPNYQHSCNFMDLNNDQQWKVGCLQNVEFNITSTCLNDSDDNSKHD |
| Parental | MDLDGSENPLALLMANGSTKRMKSVSKSRRAKIAKKVDSARLVAEQVMGDEFDLDSDDELQIDERLGKEK |
| ....G.E....LLMANG.TKR.KS.S.S..AKI.KK.D..RL..EQ.M.D.FDLD..DELQI...LGKEK | |
| Retrocopy | KAFNGNESLVPLLMANGNTKREKSLSESWQAKITKKEDTSRLMKEQAMVDTFDLDLNDELQIEKGLGKEK |
| Parental | ANLLIRSKFPRKLPRAKPCSDPNRIREPGEVEFDIEEDYTTDEDMVEGVESKLGNGSGAGGILDLLKASR |
| ..L..R.K.P..LP..K.CSD.N....PGE.E..I.E.Y..D..MVE.VE.K..NGS..GG..D.LKA.. | |
| Retrocopy | VSLVVRPKIPPSLPHVKLCSDQNQSHDPGETEITIVENYKIDKEMVEEVEGKFKNGSESGGTIDPLKAKK |
| Parental | QVGGPDYAALTEAPASPSTQEAIQGMLCMANLQSSSSSPATSSLQAWWTGGQERSSGSSSSGLGTVSSSP |
| QVGGP....LT..P.S.S....IQ...CM..LQSS..S.A.SSLQA.W.GGQ.....S.SS.LGTVS.SP | |
| Retrocopy | QVGGPGCVILTDDPNSSSNKKDIQDTVCMSDLQSSPPSQAPSSLQAFWSGGQDQGNKSPSSVLGTVSDSP |
| Parental | ASQRTPGKRPIKRPAYWKNESEEEENASLDEQDSLGACFKDAEYIYPSLESDDDD |
| .S.RTP.K.PIKR.AYW.NES.E..NA..DEQ.SLGACFK.A.YI..SLESDDDD | |
| Retrocopy | VSRRTPRKWPIKRLAYWRNES-EAGNACVDEQGSLGACFKEAKYIFLSLESDDDD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 34 .37 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 46 .63 RPM |
| SRP007412_heart | 0 .00 RPM | 20 .33 RPM |
| SRP007412_kidney | 0 .00 RPM | 29 .14 RPM |
| SRP007412_liver | 0 .00 RPM | 25 .45 RPM |
| SRP007412_testis | 30 .05 RPM | 46 .56 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_106 was not detected
No EST(s) were mapped for retro_mmus_106 retrocopy.
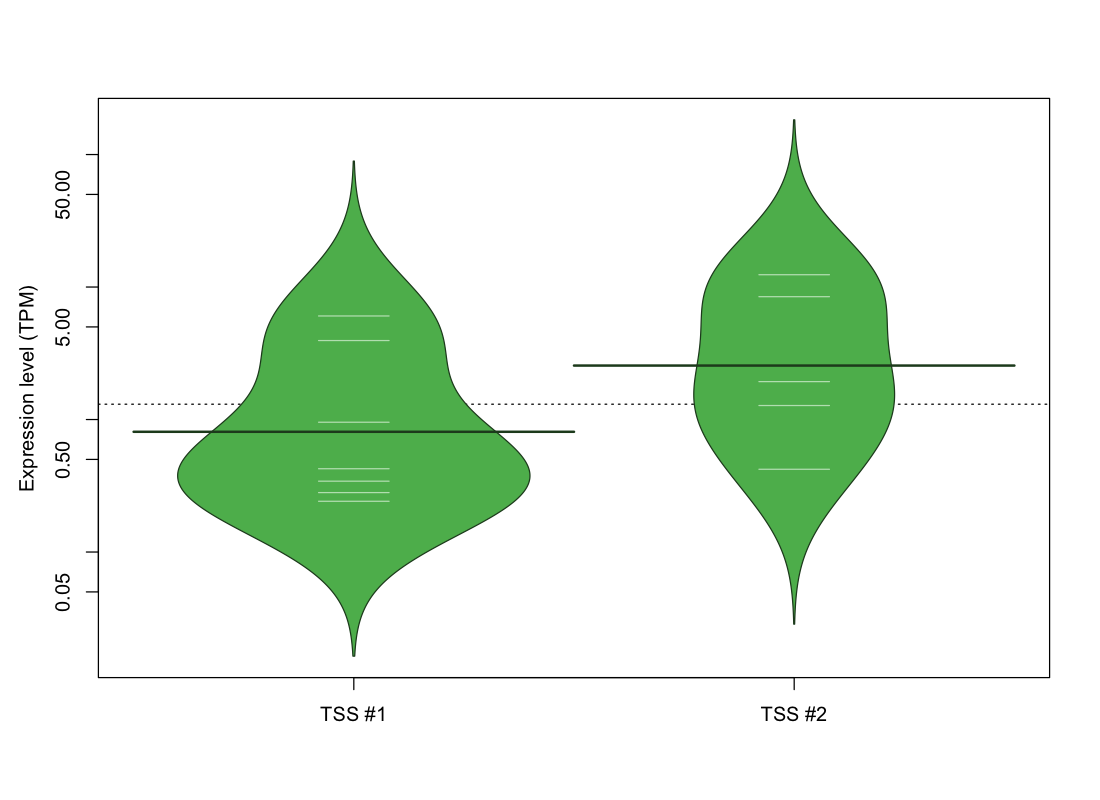
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_52459 | 1065 libraries | 5 libraries | 1 library | 1 library | 0 libraries |
| TSS #2 | TSS_52460 | 1067 libraries | 1 library | 2 libraries | 1 library | 1 library |
The graphical summary, for retro_mmus_106 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_106 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_106 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 2 parental genes, and 2 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Mus musculus | ENSMUSG00000041229 | 1 retrocopy |
retro_mmus_106 ,
|
| Rattus norvegicus | ENSRNOG00000002708 | 1 retrocopy |