RetrogeneDB ID: | retro_hsap_2887 | ||
Retrocopy location | Organism: | Human (Homo sapiens) | |
| Coordinates: | 3:178977538..178979061(-) | ||
| Located in intron of: | ENSG00000171121 | ||
Retrocopy information | Ensembl ID: | ENSG00000240429 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | LRRFIP1 | ||
| Ensembl ID: | ENSG00000124831 | ||
| Aliases: | LRRFIP1, FLAP-1, FLAP1, FLIIAP1, GCF-2, GCF2, HUFI-1, TRIP | ||
| Description: | leucine rich repeat (in FLII) interacting protein 1 [Source:HGNC Symbol;Acc:6702] |

Retrocopy-Parental alignment summary:
>retro_hsap_2887
TTGACCAGCCCCGCGGCCACGCAGAGCCGGGAGATCTACTGTTTGAGCGCGGAAGCGCAGAGGCTGGCGGAGGCCCGGCT
CGCCGCAAAACGGGAGGCCCGCGCGGAGGCTCGTGAGATCCCCATGAAGGAGCTGGAGCGGCAGCAGAAGGAGGTAGAAG
AGAGACCAGAAAAATATTTTACTGAGAAGGGGTCTCGTAACATGCTGGGCCCGTCTGCAGCCACGCTGGCCTTTCTGGGT
GGGACTTCCTCTCAGAGAGGCAGCGGAGACACCTCTATATCCATCGACACCGAGGCGTCCATCAGGGAAATCAAGGACTC
TCTAGCAGAAGTTGAAGAGAAATGTAAGAAGGCTATGTTTTCCAATGCTCAGTTAGACAATGAAAACACAAACTTCATTT
ACCAAGTTGACACCCTGAAAGATATGTTGCTGGAGATTGAAGAACAGCTGGCTGAATATAGGCGGCAGTACGAAGAGAAA
AACAAATAATTTGAAAGGGAAAAACACGCCCACAGTATACTGCAGTTTCAGTTTGCTGAAGTCAAGGAGGCCCTGAAGCA
AACAGAGGAAATGCTCGAGAAACATGGAATAATCCTAAATTCAGAAATAGTTACCAATGGAGAGACTTCCGACACTCTCA
GTAATGTTTGATACCAAGATCCTACCAAGATGACGAAAGAAGAGTTAAATGCCCTCAAGTCGACAGGGGATGGGACCCTA
GGAAAGCCAGTGAGGTGGAGGTGAAGAATGAAATCGTGGCGAATGTGGGGAAAAGAGAAATCTTGCACAATACTGAGAAA
GAACAACACACAGAGGACACAGTGAAGGATTGTGTGGACATAGAGGTATTCACTGCTGGTGAGAATACCGAGGACCAGAA
ATCCTCTGAAGACACTGCCCCATTCCTAGGAACCTTAGCAGGTGCTACCTATGAGGAACAGGTTCAAAGCCAAATTCTTG
AGAGCGCTTCTCTCCCTGAAAACACAGCACAGGTTGAGTCAAATGAGGTCATGGGTGCACCAGATGACAGGACCAGAACT
CCCCTTGAGCCATCCAACTGTTGGAGTGACTTAGATGGTGGGAGCCACACAGAGAATGTGGGAGAGGCAGCGGTGACTCA
GGTTGGAGAGCAGGCAGACACAGTGGCCTCATGTCCTTTAGGGCATAGTGATGACACAGTTTATCATGATGACAGATGTA
TGGTAGAGGTCCCCCAACAGTTAGAGACAAGCATAGGGCATAGTTTAGAGAAAGAATTCACCAACCAGGAAGCAGCTGAG
CCCAAGGAGGTTCCAGTGCAGAGTACAGAAGCAGGTAGGGATCACAACGAAGAAGAGGGTGAAGAAAAAGGATTAAGGGA
TGAGAAACCAATCAAGACAGAAGTTCCTGGTTCTCCAGCAGGAACTGAGAGCAAGGGTCAGGAGGCGACAGGTCCAAGTA
CAGTAGACACTCAAAGTGAACCCTCAGATATGAAAGAGCCAGATGAAGAAAAGAATGACCAACAGGGAGAGGCATTGGAC
TCA
ORF - retro_hsap_2887 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 91.16 % |
| Parental protein coverage: | 67.55 % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected: | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | MTSPAAAQSREIDCLSPEAQKLAEARLAAKRAARAEAREIRMKELERQQKEVEERPEKDFTEKGSRNMPG |
| .TSPAA.QSREI.CLS.EAQ.LAEARLAAKR.ARAEAREI.MKELERQQKEVEERPEK.FTEKGSRNM.G | |
| Retrocopy | LTSPAATQSREIYCLSAEAQRLAEARLAAKREARAEAREIPMKELERQQKEVEERPEKYFTEKGSRNMLG |
| Parental | LSAATLASLGGTSSRRGSGDTSISIDTEASIREIKDSLAEVEEKYKKAMVSNAQLDNEKTNFMYQVDTLK |
| .SAATLA.LGGTSS.RGSGDTSISIDTEASIREIKDSLAEVEEK.KKAM.SNAQLDNE.TNF.YQVDTLK | |
| Retrocopy | PSAATLAFLGGTSSQRGSGDTSISIDTEASIREIKDSLAEVEEKCKKAMFSNAQLDNENTNFIYQVDTLK |
| Parental | DMLLELEEQLAESRRQYEEKNKEFEREKHAHSILQFQFAEVKEALKQREEMLEKHGIILNSEIATNGETS |
| DMLLE.EEQLAE.RRQYEEKNK.FEREKHAHSILQFQFAEVKEALKQ.EEMLEKHGIILNSEI.TNGETS | |
| Retrocopy | DMLLEIEEQLAEYRRQYEEKNK*FEREKHAHSILQFQFAEVKEALKQTEEMLEKHGIILNSEIVTNGETS |
| Parental | DTLNNVGYQGPTKMTKEELNALKSTGDGTLGR-ASEVEVKNEIVANVGKREILHNTEKEQHTEDTVKDCV |
| DTL.NV.YQ.PTKMTKEELNALKSTGDGTLG..ASEVEVKNEIVANVGKREILHNTEKEQHTEDTVKDCV | |
| Retrocopy | DTLSNV*YQDPTKMTKEELNALKSTGDGTLGK<ASEVEVKNEIVANVGKREILHNTEKEQHTEDTVKDCV |
| Parental | DIEVFPAGENTEDQKSSEDTAPFLGTLAGATYEEQVQSQILESSSLPENTVQVESNEVMGAPDDRTRTPL |
| DIEVF.AGENTEDQKSSEDTAPFLGTLAGATYEEQVQSQILES.SLPENT.QVESNEVMGAPDDRTRTPL | |
| Retrocopy | DIEVFTAGENTEDQKSSEDTAPFLGTLAGATYEEQVQSQILESASLPENTAQVESNEVMGAPDDRTRTPL |
| Parental | EPSNCWSDLDGGNHTENVGEAAVTQVEEQAGTVASCPLGHSDDTVYHDDKCMVEVPQELETSTGHSLEKE |
| EPSNCWSDLDGG.HTENVGEAAVTQV.EQA.TVASCPLGHSDDTVYHDD.CMVEVPQ.LETS.GHSLEKE | |
| Retrocopy | EPSNCWSDLDGGSHTENVGEAAVTQVGEQADTVASCPLGHSDDTVYHDDRCMVEVPQQLETSIGHSLEKE |
| Parental | FTNQEAAEPKEVPAHSTEVGRDHNEEEGEETGLRDEKPIKTEVPGSPAGTEGNCQEATGPSTVDTQNEPL |
| FTNQEAAEPKEVP..STE.GRDHNEEEGEE.GLRDEKPIKTEVPGSPAGTE...QEATGPSTVDTQ.EP. | |
| Retrocopy | FTNQEAAEPKEVPVQSTEAGRDHNEEEGEEKGLRDEKPIKTEVPGSPAGTESKGQEATGPSTVDTQSEPS |
| Parental | DMKEPDEEKSDQQGEALDS |
| DMKEPDEEK.DQQGEALDS | |
| Retrocopy | DMKEPDEEKNDQQGEALDS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .44 RPM | 132 .51 RPM |
| bodymap2_adrenal | 1 .51 RPM | 161 .54 RPM |
| bodymap2_brain | 0 .80 RPM | 58 .98 RPM |
| bodymap2_breast | 0 .45 RPM | 106 .42 RPM |
| bodymap2_colon | 1 .01 RPM | 255 .16 RPM |
| bodymap2_heart | 0 .39 RPM | 115 .53 RPM |
| bodymap2_kidney | 0 .74 RPM | 191 .46 RPM |
| bodymap2_liver | 0 .38 RPM | 57 .44 RPM |
| bodymap2_lung | 0 .12 RPM | 205 .85 RPM |
| bodymap2_lymph_node | 1 .61 RPM | 133 .87 RPM |
| bodymap2_ovary | 0 .67 RPM | 97 .39 RPM |
| bodymap2_prostate | 1 .35 RPM | 244 .72 RPM |
| bodymap2_skeletal_muscle | 0 .55 RPM | 95 .05 RPM |
| bodymap2_testis | 1 .51 RPM | 80 .48 RPM |
| bodymap2_thyroid | 1 .22 RPM | 143 .72 RPM |
| bodymap2_white_blood_cells | 0 .18 RPM | 428 .76 RPM |
RNA Polymerase II activity may be related with retro_hsap_2887 in 31 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CFW | POLR2A | 3:178979022..178979365 |
| ENCFF002CFX | POLR2A | 3:178979023..178979349 |
| ENCFF002CGN | POLR2A | 3:178979039..178979340 |
| ENCFF002CHO | POLR2A | 3:178978909..178979402 |
| ENCFF002CIO | POLR2A | 3:178979039..178979260 |
| ENCFF002CJZ | POLR2A | 3:178979085..178979301 |
| ENCFF002CKX | POLR2A | 3:178979027..178979299 |
| ENCFF002CLM | POLR2A | 3:178978935..178979475 |
| ENCFF002CMI | POLR2A | 3:178978864..178979410 |
| ENCFF002COJ | POLR2A | 3:178979010..178979446 |
| ENCFF002CQC | POLR2A | 3:178979024..178979321 |
| ENCFF002CQE | POLR2A | 3:178979126..178979339 |
| ENCFF002CQG | POLR2A | 3:178978945..178979469 |
| ENCFF002CQI | POLR2A | 3:178979091..178979231 |
| ENCFF002CQK | POLR2A | 3:178979007..178979326 |
| ENCFF002CQM | POLR2A | 3:178979030..178979307 |
| ENCFF002CQO | POLR2A | 3:178979040..178979296 |
| ENCFF002CSY | POLR2A | 3:178979013..178979313 |
| ENCFF002CUP | POLR2A | 3:178979026..178979342 |
| ENCFF002CXM | POLR2A | 3:178978945..178979435 |
| ENCFF002CXO | POLR2A | 3:178978980..178979404 |
| ENCFF002CXP | POLR2A | 3:178979145..178979273 |
| ENCFF002CXR | POLR2A | 3:178979075..178979297 |
| ENCFF002DAH | POLR2A | 3:178979040..178979324 |
| ENCFF002DBB | POLR2A | 3:178979056..178979256 |
| ENCFF002DBE | POLR2A | 3:178979104..178979285 |
| ENCFF002DBO | POLR2A | 3:178979059..178979301 |
| ENCFF002DBP | POLR2A | 3:178979061..178979286 |
| ENCFF002DBQ | POLR2A | 3:178979011..178979381 |
| ENCFF002DBT | POLR2A | 3:178978998..178979368 |
| ENCFF002CZW | POLR2A | 3:178978730..178979480 |
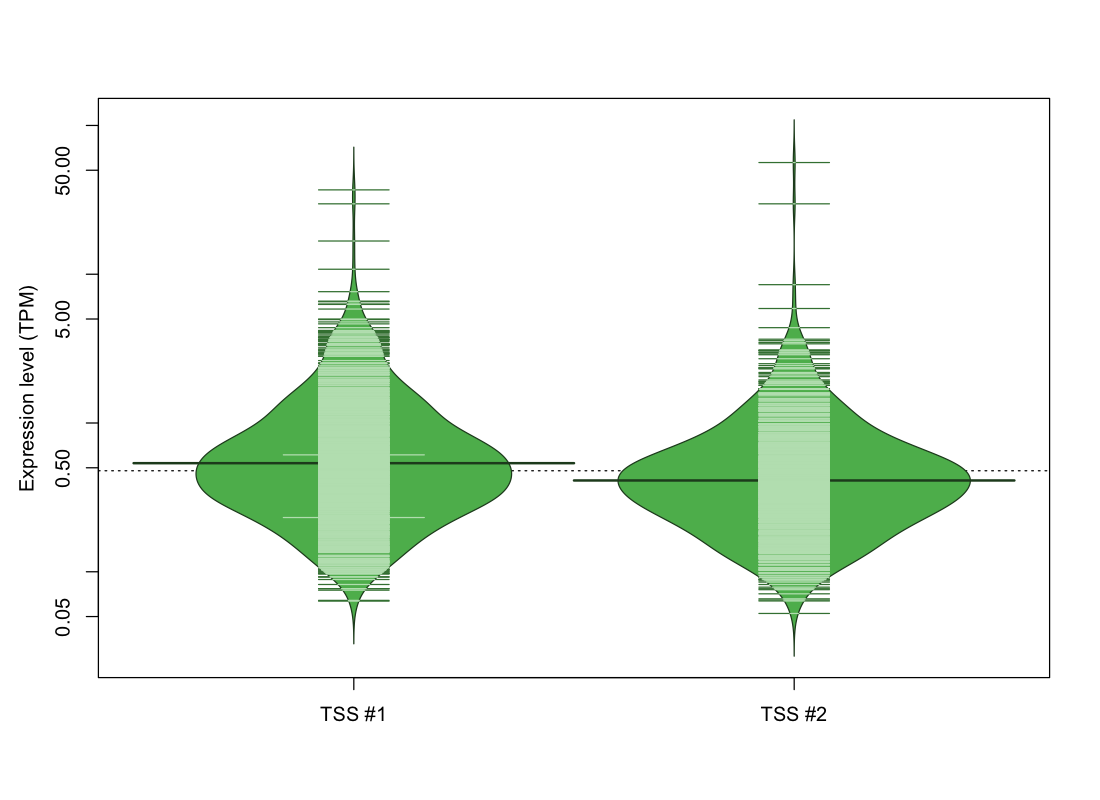
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_131962 | 931 libraries | 697 libraries | 191 libraries | 6 libraries | 4 libraries |
| TSS #2 | TSS_131963 | 1128 libraries | 611 libraries | 86 libraries | 2 libraries | 2 libraries |
The graphical summary, for retro_hsap_2887 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_2887 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2887 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_2001 |
| Macaca mulatta | retro_mmul_1455 |
Parental genes homology:
Parental genes homology involve 3 parental genes, and 3 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000124831 | 1 retrocopy |
retro_hsap_2887 ,
|
| Gorilla gorilla | ENSGGOG00000008411 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000003386 | 1 retrocopy |
Expression level across human populations :
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Undefined variable $populLEGEND in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3353
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .00 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .00 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .00 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .00 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .00 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .00 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .00 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .00 RPM |
Fatal error: Uncaught Error: Call to undefined function mysql_query() in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php:3390 Stack trace: #0 /home/rosikiewicz/RetrogeneDB2/BETA/retrogene.php(1158): include() #1 {main} thrown in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3390