RetrogeneDB ID: | retro_hsap_4123 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 9:5110913..5112109(+) | ||
| Located in intron of: | ENSG00000096968 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000236567 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | TCF3 | ||
| Ensembl ID: | ENSG00000071564 | ||
| Aliases: | TCF3, E2A, E47, ITF1, TCF-3, VDIR, bHLHb21 | ||
| Description: | transcription factor 3 [Source:HGNC Symbol;Acc:11633] |

Retrocopy-Parental alignment summary:
>retro_hsap_4123
ATGAACCAGCCGCAGAGGATGGCATCCGTGGACACGGACAAGGAGCTCAGTAACCTGGACTTCAGCATGATGTTCCCGCT
GCCCGTTGCCAACGGGAAGGGCCGGCCCGCCTCCCTGGCCGGGGCGCAATTCAGAGGTTCAGGTCTTGAGAACTGGCCCA
GCTCAGGCTCCTGGGGCAGCGGCGACCAGAACAGCTCCTCCTTTGACCCCAGCTGGACGTTCAGCGAAGGCGCTCAACTT
GCTGAGTCGCACGGCAGCCACTCTCCATTCACATTCCTGGGACCGGGACTCGGAGGCAAGAGCAGCTAGCGCGGGCCTAT
TCCTCCTTTGGTAGAGACGCAGGCGTGCGCAGCCTGACTCAGGCTGGCTTCCTGCCGGGCAAGCTGGCCCTCAGCAGCCC
CGGACCCCTGTCCCCCTCAGGCATGAAGGGGACCTCCCGGTACTACCCCTCCTACGCCAGCAGCTCTGGCGGACAGAGGC
GGACAGCGGCCTGGACACGCAGCCCACGAAGGTCGGGAAGGTCCCGCCTGGTCTTCCATCCTCGGCGTACCTGCCCAGCT
CAGGTGAGGAGTACGGTAGGGATGCCGCCGCCTATCCATCCGCCAAGACGCCCAGCAGCGCCTGTCCCGCCCCCTTCTAC
ATGGCAGATGGCAGCCTGCACCCCTAAGCCGAGCTCTGGAGTTCCCTGGGCCAGGTGGGCTTTGGCCCCATGCTGGGCGG
GGGCTCATGCCCCTCGTCCCTCCCGCCCAGCAGCGGCCCTGTGGGCAGTGGCGGAGGCAGCAGCACGTTTGGCGGCCTGC
ACCAGCACGAGCGCGTGGGCTACCGGCTGCACGGAGGAGAAGTGAACGGTGGGCTTCCGGCTGCATCCACCTTCTCCTTG
GCCCCCGGAGCCACGTAGGCGGCGTCTCCAGCCACACGCCTGTCGGCGGGGCCGACAACCTCCTGGGCTCCCGAGGGACC
ACAGCCAGCAGCTCTGGGGACGCCCTCGGGAAGGCCTGGCCTCAATCTACTCTCCGGATCACTCAAGCAATAACTTGTGG
TCCAGCCCCTCCACCGCCGTGGGCTCCCCCCAGGGCCTGGGAGGGACGTCGCACTAGCCTGGAGCAGGAGTCCCTGGTGC
CTTATCACCCAGCTACGACGGGGGTCTCCACGGCCTGGAGAGTAAGATAGAAGACCACCTACCTGAACGAGGGCAT
ATGAACCAGCCGCAGAGGATGGCATCCGTGGACACGGACAAGGAGCTCAGTAACCTGGACTTCAGCATGATGTTCCCGCT
GCCCGTTGCCAACGGGAAGGGCCGGCCCGCCTCCCTGGCCGGGGCGCAATTCAGAGGTTCAGGTCTTGAGAACTGGCCCA
GCTCAGGCTCCTGGGGCAGCGGCGACCAGAACAGCTCCTCCTTTGACCCCAGCTGGACGTTCAGCGAAGGCGCTCAACTT
GCTGAGTCGCACGGCAGCCACTCTCCATTCACATTCCTGGGACCGGGACTCGGAGGCAAGAGCAGCTAGCGCGGGCCTAT
TCCTCCTTTGGTAGAGACGCAGGCGTGCGCAGCCTGACTCAGGCTGGCTTCCTGCCGGGCAAGCTGGCCCTCAGCAGCCC
CGGACCCCTGTCCCCCTCAGGCATGAAGGGGACCTCCCGGTACTACCCCTCCTACGCCAGCAGCTCTGGCGGACAGAGGC
GGACAGCGGCCTGGACACGCAGCCCACGAAGGTCGGGAAGGTCCCGCCTGGTCTTCCATCCTCGGCGTACCTGCCCAGCT
CAGGTGAGGAGTACGGTAGGGATGCCGCCGCCTATCCATCCGCCAAGACGCCCAGCAGCGCCTGTCCCGCCCCCTTCTAC
ATGGCAGATGGCAGCCTGCACCCCTAAGCCGAGCTCTGGAGTTCCCTGGGCCAGGTGGGCTTTGGCCCCATGCTGGGCGG
GGGCTCATGCCCCTCGTCCCTCCCGCCCAGCAGCGGCCCTGTGGGCAGTGGCGGAGGCAGCAGCACGTTTGGCGGCCTGC
ACCAGCACGAGCGCGTGGGCTACCGGCTGCACGGAGGAGAAGTGAACGGTGGGCTTCCGGCTGCATCCACCTTCTCCTTG
GCCCCCGGAGCCACGTAGGCGGCGTCTCCAGCCACACGCCTGTCGGCGGGGCCGACAACCTCCTGGGCTCCCGAGGGACC
ACAGCCAGCAGCTCTGGGGACGCCCTCGGGAAGGCCTGGCCTCAATCTACTCTCCGGATCACTCAAGCAATAACTTGTGG
TCCAGCCCCTCCACCGCCGTGGGCTCCCCCCAGGGCCTGGGAGGGACGTCGCACTAGCCTGGAGCAGGAGTCCCTGGTGC
CTTATCACCCAGCTACGACGGGGGTCTCCACGGCCTGGAGAGTAAGATAGAAGACCACCTACCTGAACGAGGGCAT
ORF - retro_hsap_4123 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 80.3 % |
| Parental protein coverage: | 61.75 % |
| Number of stop codons detected: | 3 |
| Number of frameshifts detected | 4 |
Retrocopy - Parental Gene Alignment:
| Parental | MNQPQRMAPVGTDKELSDLLDFSMMFPLPVTNGKGRPASLAGAQFGGSGLEDRPSSGSWGSGDQSSSSFD |
| MNQPQRMA.V.TDKELS.L.DFSMMFPLPV.NGKGRPASLAGAQF.GSGLE..PSSGSWGSGDQ.SSSFD | |
| Retrocopy | MNQPQRMASVDTDKELSNL-DFSMMFPLPVANGKGRPASLAGAQFRGSGLENWPSSGSWGSGDQNSSSFD |
| Parental | PSRTFSEGTHFTESHSSLSSSTFLGPGLGGKSGERG-AYASFGRDAGVGGLTQAGFLSGELALNSPGPLS |
| PS.TFSEG....ESH.S.S..TFLGPGLGGKS..RG.AY.SFGRDAGV..LTQAGFL.G.LAL.SPGPLS | |
| Retrocopy | PSWTFSEGAQLAESHGSHSPFTFLGPGLGGKSS*RG<AYSSFGRDAGVRSLTQAGFLPGKLALSSPGPLS |
| Parental | PSGMKGTSQYYPSYSGSSR-RRAADGSLDTQPKKVRKVPPGLPSSVYPPSSGEDYGRDATAYPSAKTPSS |
| PSGMKGTS.YYPSY..SS..R..AD..LDTQP.KV.KVPPGLPSS.Y.PSSGE.YGRDA.AYPSAKTPSS | |
| Retrocopy | PSGMKGTSRYYPSYASSSG<RTEADSGLDTQPTKVGKVPPGLPSSAYLPSSGEEYGRDAAAYPSAKTPSS |
| Parental | TYPAPFYVADGSLHPSAELWSPPGQAGFGPMLGGGSSPLPLPPGSGPVGSSGSSSTFGGLHQHERMGYQL |
| ..PAPFY.ADGSLHP.AELWS..GQ.GFGPMLGGGS.P..LPP.SGPVGS.G.SSTFGGLHQHER.GY.L | |
| Retrocopy | ACPAPFYMADGSLHP*AELWSSLGQVGFGPMLGGGSCPSSLPPSSGPVGSGGGSSTFGGLHQHERVGYRL |
| Parental | HGAEVNGGLPSASSFSSAPGAT-YGGVSSHTPPVSGADSLLGSRGTTAGSSGDALGKA-LASIYSPDHSS |
| HG.EVNGGLP.AS.FS.APGAT..GGVSSHT.PV.GAD.LLGSRGTTA.SSGDALGKA.LASIYSPDHSS | |
| Retrocopy | HGGEVNGGLPAASTFSLAPGAT<VGGVSSHT-PVGGADNLLGSRGTTASSSGDALGKA<LASIYSPDHSS |
| Parental | NNFSSSPSTPVGSPQGLAGTSQWPRAGAPGALSPSYDGGLHGLQSKIEDHLDEAIH |
| NN..SSPST.VGSPQGL.GTS..P.AG.PGALSPSYDGGLHGL.SKIEDHL.E..H | |
| Retrocopy | NNLWSSPSTAVGSPQGLGGTSH*PGAGVPGALSPSYDGGLHGLESKIEDHLPERGH |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .02 RPM | 23 .10 RPM |
| bodymap2_adrenal | 0 .10 RPM | 47 .18 RPM |
| bodymap2_brain | 0 .00 RPM | 5 .88 RPM |
| bodymap2_breast | 0 .00 RPM | 21 .36 RPM |
| bodymap2_colon | 0 .00 RPM | 25 .14 RPM |
| bodymap2_heart | 0 .11 RPM | 8 .20 RPM |
| bodymap2_kidney | 0 .04 RPM | 14 .72 RPM |
| bodymap2_liver | 0 .00 RPM | 4 .96 RPM |
| bodymap2_lung | 0 .04 RPM | 21 .48 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 76 .49 RPM |
| bodymap2_ovary | 0 .04 RPM | 51 .95 RPM |
| bodymap2_prostate | 0 .00 RPM | 26 .05 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 13 .88 RPM |
| bodymap2_testis | 0 .08 RPM | 141 .38 RPM |
| bodymap2_thyroid | 0 .00 RPM | 24 .95 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 35 .75 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_4123 was not detected
No EST(s) were mapped for retro_hsap_4123 retrocopy.
No TSS is located nearby retro_hsap_4123 retrocopy 5' end.
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | CTCTCCGGATCACTCAAGCA (20 nt long) |
| Reverse primer: | GACTCCTGCTCCAGGCTAGT (20 nt long) |
| Anneling temperature: | 60 °C |
| (Expected) product size: | 103 |
| Electrophoresis gel image: | 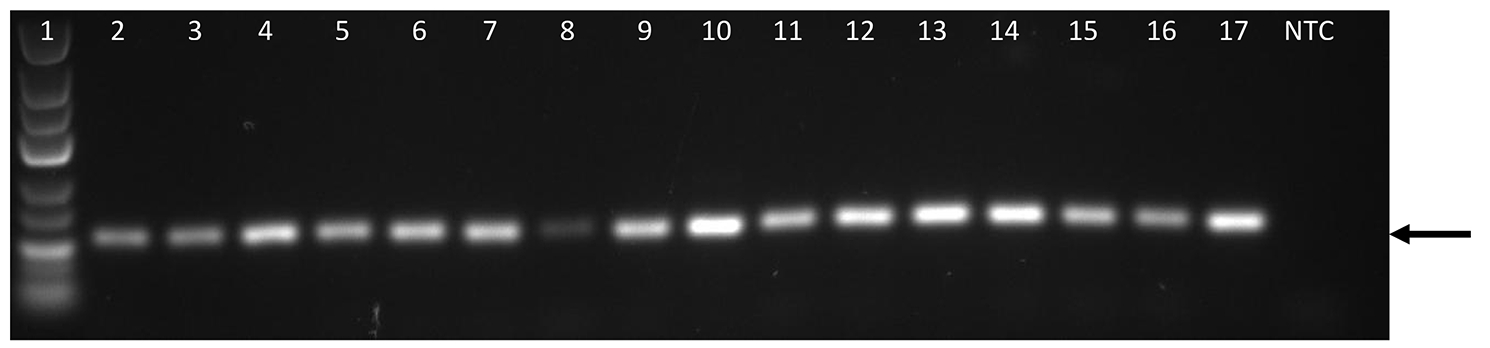 |
| Additional comment: | Black arrows indicate PCR products of retrogenes undergoing deletions in various human populations (A-J). Lane 1 –GeneRuler Low Range DNA Ladder (Thermo Scientific); 2 –Heart; 3 –Brain; 4 –Placenta; 5 –Lung; 6 –Liver; 7 –Skeletal Muscle; 8 –Kidney; 9 –Pancreas; 10 –Spleen; 11 –Thymus; 12 –Prostate; 13 –Testis; 14 –Ovary, 15 –Small intestine 16 –Colon; 17 –Leukocyte; NTC—No template control (water instead of cDNA). |
Retrocopy orthology:
Retrocopy retro_hsap_4123 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_2770 |
| Macaca mulatta | retro_mmul_1194 |
Parental genes homology:
Parental genes homology involve 4 parental genes, and 4 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000071564 | 1 retrocopy |
retro_hsap_4123 ,
|
| Gorilla gorilla | ENSGGOG00000003667 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000009017 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000001742 | 1 retrocopy |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .06 RPM |
| CEU_NA11930 | 0 .07 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .07 RPM |
| CEU_NA12751 | 0 .05 RPM |
| CEU_NA12760 | 0 .04 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .05 RPM |
| CEU_NA12873 | 0 .03 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .07 RPM |
| FIN_HG00315 | 0 .00 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .12 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .03 RPM |
| FIN_HG00375 | 0 .02 RPM |
| FIN_HG00377 | 0 .03 RPM |
| FIN_HG00378 | 0 .02 RPM |
| GBR_HG00099 | 0 .03 RPM |
| GBR_HG00111 | 0 .02 RPM |
| GBR_HG00114 | 0 .08 RPM |
| GBR_HG00119 | 0 .02 RPM |
| GBR_HG00131 | 0 .06 RPM |
| GBR_HG00133 | 0 .10 RPM |
| GBR_HG00134 | 0 .09 RPM |
| GBR_HG00137 | 0 .08 RPM |
| GBR_HG00142 | 0 .11 RPM |
| GBR_HG00143 | 0 .03 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .10 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .03 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .08 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .07 RPM |
| YRI_NA18907 | 0 .03 RPM |
| YRI_NA18916 | 0 .06 RPM |
| YRI_NA19093 | 0 .03 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .04 RPM |
| YRI_NA19213 | 0 .05 RPM |
| YRI_NA19214 | 0 .12 RPM |
| YRI_NA19223 | 0 .08 RPM |
Indel association:
The presence of retro_hsap_4123 across human populations is associated with 1 indel. The percentage values indicate the frequencies of retro_hsap_4123 presence in various populations. Based on Kabza et al. 2015 (PubMed).| # | Indel coordinates | AFR, African | AMR, Ad Mixed American | EUR, European | EAS, East Asian | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASW | YRI | LWK | MXL | PUR | CLM | CEU | IBS | GBR | FIN | TSI | JPT | CHB | CHS | ||
| 1. | 9:5106619..5115630 | 99.18 | 99.43 | 98.97 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
Indel #1, located at the genomic coordinates 9:5106619..5115630.