RetrogeneDB ID: | retro_hsap_4862 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | X:74963062..74966746(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000215105 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | TTC3 | ||
| Ensembl ID: | ENSG00000182670 | ||
| Aliases: | TTC3, DCRR1, RNF105, TPRDIII | ||
| Description: | tetratricopeptide repeat domain 3 [Source:HGNC Symbol;Acc:12393] |

Retrocopy-Parental alignment summary:
>retro_hsap_4862
GACAATTTTGCTGAGGGAGATTTCACTATGGCGGATTATGCCTTGTTAGAAGATTGCCCTTACGTGGACGATTGTGTCTT
TGCTGCTGAATTTATGACCGATGATTATGTTCGTGTGACTCAGCTTTACTGTGATGGGGTGGGTAAGCAATATAAAGATT
ATGTCCAAAGTGAGAGGAATTTAGAATTTGACATCTGCAGTATATGGTGTAGTAAACCAATTTCTGTCCTGCAAGATTAT
TGTGATGCCATTAAAATAAACATCTTCTGGCCACTTCTGTTTCAACATCAAAACAGTTCTGTAATATCACGATTGCATCC
CTGTGTGGACGCCAACAATTCACGTGCTTCCGAGATAAATTTGAAGAGATTACAACATCTTGAGTTGATGGAAGATATTG
TGGATTTGGCAAAGAAAGCTGCTAATGATTCACTCCTTATTGGAGGCTTATTGAGAATTGGTTATAAAATAGAAAATAAA
ATCTTGGCAATGGAAGAAGCTCTGAATTGGATAAAATATGCAGGCGATGTAACAATTCTAACTAAATTAGGATCAATTGA
CAATTGTTGGCCTATGTTAAGTATTTTCTTTACTGAATACAAGTACCACATAACTAAAATTGTAATGGAAGACTGCAATT
TGCTTGAAGAACTTAAAACCCAAAGTTGTATGGATTGTATAGAGCAAGGAGAACTAATGAAAATGAAAGGAAATGAAGAG
TTTTCCAAAGAAAGATTTGATATAGCTATTATCTATTACACCAGAGCCATTGAATATAGACCTGAAAACCACCTTCTTTA
TGGTAACCGAGCTCTTTGTTTTCTTCGTACTGGACAGTTTAGAAATGCACTTGGTGATGGAAAGAGAGCCACTATTCTGA
AGAACACTTGGCCAAAGGGTCATTATCGTTATTGTGATGCTCTTTCTATGCTGGGGGAATATAACTGGGCCCTGCAAGCA
AACATAACAGCTCAAAAACTCTGTAAAAATGACCCTGAGGGAATCAAGGATCTAATTCAGCAGCATGTAAAGTTACAAAA
ACAAATAGAAGACCTACAAGGTCGAACAGCAAATAAGAATCCAATTAAAGCCTTTTATGAAAACAGGGCCTACACACCTA
GGAGTTTATCAGCACCTGTATTTAGTACTTCACTTAACTTTGTGGAGAAGGAAAGAGATTTCAGAAAAATTAATCACGAA
ATGGCTAACGGTGGTAATCAGAATCTAAAGGTGATGGATGAGGCATTGATGGTAGATGATTGTGACTGTCATCCTGAATT
TTCACCACCATCAAGTCAGCCTCCGAAACATAAAGGAAAACAAAAATCTCGAAACAATGAATCAGAAAAGTTCAGTTCTA
GTTCACAATTGACTTTACCAGCAGATTTGAAGAACATCTTGGAGAAACAGTTTTCTAAATCTTCCAGAGGTGCACACCAG
GATTTTGCTAATATAATGACAATGCTGAGAAGCTTAATTCAAGATGGCTATACAGCCTTATTGGAGCAGCGTTGCCGCAG
TGCCGCACATGCCTTTACAGAGTTGCTGAATGGTTTAGATCCTCAAAAAATAAAGCAATTGAATCTGGCCATGATTAACT
ATGTCTTGGTTGTCTATGGACTTGCCATTTCACTCCTTGGAATAGGACAGCCTGAAGAACTATCTGAAGCTGAAAACCAG
TTTAAGAGGATTATTGAACACTACCACAATGAGGGACTTGATTGCTTGGCCTACTGTGGAATTGGAAAAGTATATTTGAA
AAAAAACAGATTTCTAGAAGCTCTCAATCACTTTGAGAAAGCAAGAACCTTGATTTATCGTCTTCCTGGAGTGTTAACTT
GGCCCACAAGTAATGTGATTATTGAAGAGTCTCAGCCAGAAAAAATAAAGATGCTGTTAGAGAAATTTGTTGAAGAATGC
AAGTTCCCTCCAGTGCCAGATGCCATTTGTTGCTATCAGAAGTGCTGTGGATATTCTAAGATCCAGATATACATAACTGA
TCCAGACTTTAAGGGTTTTATACGCATCAGCTGTTGCCAGTACTGTAAAATAGAATTTCACATGAATTGCTGGAAGAAGT
TAAAAACCACAACCTTTAATGATAAAATTGACAAGGATTTTCTACAAGGAATATGTCTTACCCCTGACTGTGAAGGTGTC
ATTTCTAAGATTATCATCTTCAGCAGTGGTGGTCAAGTTAAATGTGAATTTGAACACAAGGTCATAAAAGAAAAGGTTCC
TCCAAGACCTATTCTGAAACAGAAATGTTCTAGCCTAGAGAAACTAAGACTGAAAGAAGACAAAAAATTGAAGAGAAAGA
TCCAAAAAAAAGAAGCAAAAAAATTAGCACAAGAAAGAATGGAGGAGGACTTAAGAGAAAGTAATCCACCCAAAAACGAA
GAGCAGAAAGAAACTGTAGACAATGTTCAGCATTGTCAGTTCCTTGATGACAGAATTCTACAGTGTATAAAGCAGTATGC
TGACAAGATTAAATCCGGCATATGGAATACAGCCACGCTTCTCAAAGAATTACTTTCTTGGAAAGTTTTGAGCACAGAGG
ACTATACAACCTGTTTTTCAAGCAGAAATTTTCTAAATGAAGCAGTGGACTATGTTATTCGTCACTTGATTCAAGAAAAG
AACAGAGTAAAGACAAGAATATTTCTGCATGTTTTGAGTGAGCTTAAAGAAGTGGAGCCCAAATTAGCCGCCTGGATCCG
AAAACTTAATAGCTTTGGCTTAGATGCCACAGGACCTTTCTTTTCTCGGTATGAAGCATCTCTTAAACAGCTTGATTTTA
GCATCATGACTTTCCTCTGGAATGAGAAATATGGTCACAAACTAGACTCTATAGAAGGAAAGCAACTTGATTATTTCTTT
GAGCCAACATCATCGAAGGAAGCCCGCTGTTTAATATGGCTGCTAGAAGAACACAGAGACAAGTTCCCAGCATTACATAG
TGCTTTAGATGAATTCTTTGATATAATGGACAGCCGCTGTACTGTGTTAAGGAAACAAGACAGTGGCGAAGCACCGTTTA
GTTCTACCAAGGTGAAAAACAAAGGCAAGAAAAAGAAACCAAAGGATTCAAAGCCTATGTTAGTTGGGTCTGGAACAACT
TCAGTAACTCCAAATAATGAGATCATCACTTCAAGTGAAGACCATAGCAATCAAAATTCAGATACTGCAGGCCCATTTGC
AGTGCCTGACCATCTTCGGCAAGACGTAGAAGAATTTGAAGCTCTCTATGACCAACACAGTAATGAATATGTTGTCCGCA
ATAAGAAGCTATGGGACATGAACCCAAAACAAAAATGTTCAACTCTATATGATTACTTCTCTCAGTTGTTGGAGGAACAT
GGTCCCTTGGACATGAGTCACAAGATGTTCTCTGAAGAATATGAGTTTTTCCCAGAAGAAACTCGACCAATACTAGAAAA
AGCAGGAGGTTTAAAATCTTTTCTCTTGGGATGTCCCCGTTTTGTTGTGATTGACAACTGTATTGCATTGAAGAAAGTTG
CATCACGGCTCAAGAAAAAAAGAAAGAAGAAAAACATTAAAACAAAAGTAGAAGACATTTCAAAAGCAGGAGAGTATTTA
CAAGTTAAACTACCACTTAATTCAGCTGCTAGGGAATTTAAACCAGATGTAAAGTCTAAACCAGTATCAGGTTCATCTTC
AGCA
GACAATTTTGCTGAGGGAGATTTCACTATGGCGGATTATGCCTTGTTAGAAGATTGCCCTTACGTGGACGATTGTGTCTT
TGCTGCTGAATTTATGACCGATGATTATGTTCGTGTGACTCAGCTTTACTGTGATGGGGTGGGTAAGCAATATAAAGATT
ATGTCCAAAGTGAGAGGAATTTAGAATTTGACATCTGCAGTATATGGTGTAGTAAACCAATTTCTGTCCTGCAAGATTAT
TGTGATGCCATTAAAATAAACATCTTCTGGCCACTTCTGTTTCAACATCAAAACAGTTCTGTAATATCACGATTGCATCC
CTGTGTGGACGCCAACAATTCACGTGCTTCCGAGATAAATTTGAAGAGATTACAACATCTTGAGTTGATGGAAGATATTG
TGGATTTGGCAAAGAAAGCTGCTAATGATTCACTCCTTATTGGAGGCTTATTGAGAATTGGTTATAAAATAGAAAATAAA
ATCTTGGCAATGGAAGAAGCTCTGAATTGGATAAAATATGCAGGCGATGTAACAATTCTAACTAAATTAGGATCAATTGA
CAATTGTTGGCCTATGTTAAGTATTTTCTTTACTGAATACAAGTACCACATAACTAAAATTGTAATGGAAGACTGCAATT
TGCTTGAAGAACTTAAAACCCAAAGTTGTATGGATTGTATAGAGCAAGGAGAACTAATGAAAATGAAAGGAAATGAAGAG
TTTTCCAAAGAAAGATTTGATATAGCTATTATCTATTACACCAGAGCCATTGAATATAGACCTGAAAACCACCTTCTTTA
TGGTAACCGAGCTCTTTGTTTTCTTCGTACTGGACAGTTTAGAAATGCACTTGGTGATGGAAAGAGAGCCACTATTCTGA
AGAACACTTGGCCAAAGGGTCATTATCGTTATTGTGATGCTCTTTCTATGCTGGGGGAATATAACTGGGCCCTGCAAGCA
AACATAACAGCTCAAAAACTCTGTAAAAATGACCCTGAGGGAATCAAGGATCTAATTCAGCAGCATGTAAAGTTACAAAA
ACAAATAGAAGACCTACAAGGTCGAACAGCAAATAAGAATCCAATTAAAGCCTTTTATGAAAACAGGGCCTACACACCTA
GGAGTTTATCAGCACCTGTATTTAGTACTTCACTTAACTTTGTGGAGAAGGAAAGAGATTTCAGAAAAATTAATCACGAA
ATGGCTAACGGTGGTAATCAGAATCTAAAGGTGATGGATGAGGCATTGATGGTAGATGATTGTGACTGTCATCCTGAATT
TTCACCACCATCAAGTCAGCCTCCGAAACATAAAGGAAAACAAAAATCTCGAAACAATGAATCAGAAAAGTTCAGTTCTA
GTTCACAATTGACTTTACCAGCAGATTTGAAGAACATCTTGGAGAAACAGTTTTCTAAATCTTCCAGAGGTGCACACCAG
GATTTTGCTAATATAATGACAATGCTGAGAAGCTTAATTCAAGATGGCTATACAGCCTTATTGGAGCAGCGTTGCCGCAG
TGCCGCACATGCCTTTACAGAGTTGCTGAATGGTTTAGATCCTCAAAAAATAAAGCAATTGAATCTGGCCATGATTAACT
ATGTCTTGGTTGTCTATGGACTTGCCATTTCACTCCTTGGAATAGGACAGCCTGAAGAACTATCTGAAGCTGAAAACCAG
TTTAAGAGGATTATTGAACACTACCACAATGAGGGACTTGATTGCTTGGCCTACTGTGGAATTGGAAAAGTATATTTGAA
AAAAAACAGATTTCTAGAAGCTCTCAATCACTTTGAGAAAGCAAGAACCTTGATTTATCGTCTTCCTGGAGTGTTAACTT
GGCCCACAAGTAATGTGATTATTGAAGAGTCTCAGCCAGAAAAAATAAAGATGCTGTTAGAGAAATTTGTTGAAGAATGC
AAGTTCCCTCCAGTGCCAGATGCCATTTGTTGCTATCAGAAGTGCTGTGGATATTCTAAGATCCAGATATACATAACTGA
TCCAGACTTTAAGGGTTTTATACGCATCAGCTGTTGCCAGTACTGTAAAATAGAATTTCACATGAATTGCTGGAAGAAGT
TAAAAACCACAACCTTTAATGATAAAATTGACAAGGATTTTCTACAAGGAATATGTCTTACCCCTGACTGTGAAGGTGTC
ATTTCTAAGATTATCATCTTCAGCAGTGGTGGTCAAGTTAAATGTGAATTTGAACACAAGGTCATAAAAGAAAAGGTTCC
TCCAAGACCTATTCTGAAACAGAAATGTTCTAGCCTAGAGAAACTAAGACTGAAAGAAGACAAAAAATTGAAGAGAAAGA
TCCAAAAAAAAGAAGCAAAAAAATTAGCACAAGAAAGAATGGAGGAGGACTTAAGAGAAAGTAATCCACCCAAAAACGAA
GAGCAGAAAGAAACTGTAGACAATGTTCAGCATTGTCAGTTCCTTGATGACAGAATTCTACAGTGTATAAAGCAGTATGC
TGACAAGATTAAATCCGGCATATGGAATACAGCCACGCTTCTCAAAGAATTACTTTCTTGGAAAGTTTTGAGCACAGAGG
ACTATACAACCTGTTTTTCAAGCAGAAATTTTCTAAATGAAGCAGTGGACTATGTTATTCGTCACTTGATTCAAGAAAAG
AACAGAGTAAAGACAAGAATATTTCTGCATGTTTTGAGTGAGCTTAAAGAAGTGGAGCCCAAATTAGCCGCCTGGATCCG
AAAACTTAATAGCTTTGGCTTAGATGCCACAGGACCTTTCTTTTCTCGGTATGAAGCATCTCTTAAACAGCTTGATTTTA
GCATCATGACTTTCCTCTGGAATGAGAAATATGGTCACAAACTAGACTCTATAGAAGGAAAGCAACTTGATTATTTCTTT
GAGCCAACATCATCGAAGGAAGCCCGCTGTTTAATATGGCTGCTAGAAGAACACAGAGACAAGTTCCCAGCATTACATAG
TGCTTTAGATGAATTCTTTGATATAATGGACAGCCGCTGTACTGTGTTAAGGAAACAAGACAGTGGCGAAGCACCGTTTA
GTTCTACCAAGGTGAAAAACAAAGGCAAGAAAAAGAAACCAAAGGATTCAAAGCCTATGTTAGTTGGGTCTGGAACAACT
TCAGTAACTCCAAATAATGAGATCATCACTTCAAGTGAAGACCATAGCAATCAAAATTCAGATACTGCAGGCCCATTTGC
AGTGCCTGACCATCTTCGGCAAGACGTAGAAGAATTTGAAGCTCTCTATGACCAACACAGTAATGAATATGTTGTCCGCA
ATAAGAAGCTATGGGACATGAACCCAAAACAAAAATGTTCAACTCTATATGATTACTTCTCTCAGTTGTTGGAGGAACAT
GGTCCCTTGGACATGAGTCACAAGATGTTCTCTGAAGAATATGAGTTTTTCCCAGAAGAAACTCGACCAATACTAGAAAA
AGCAGGAGGTTTAAAATCTTTTCTCTTGGGATGTCCCCGTTTTGTTGTGATTGACAACTGTATTGCATTGAAGAAAGTTG
CATCACGGCTCAAGAAAAAAAGAAAGAAGAAAAACATTAAAACAAAAGTAGAAGACATTTCAAAAGCAGGAGAGTATTTA
CAAGTTAAACTACCACTTAATTCAGCTGCTAGGGAATTTAAACCAGATGTAAAGTCTAAACCAGTATCAGGTTCATCTTC
AGCA
ORF - retro_hsap_4862 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.08 % |
| Parental protein coverage: | 60.64 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | DNFAEGDFTVADYALLEDCPHVDDCVFAAEFMSNDYVRVTQLYCDGVGVQYKDYIQSERNLEFDICSIWC |
| DNFAEGDFT.ADYALLEDCP.VDDCVFAAEFM..DYVRVTQLYCDGVG.QYKDY.QSERNLEFDICSIWC | |
| Retrocopy | DNFAEGDFTMADYALLEDCPYVDDCVFAAEFMTDDYVRVTQLYCDGVGKQYKDYVQSERNLEFDICSIWC |
| Parental | SKPISVLQDYCDAIKINIFWPLLFQHQNSSVISRLHPCVDANNSRASEINLKKLQHLELMEDIVDLAKKV |
| SKPISVLQDYCDAIKINIFWPLLFQHQNSSVISRLHPCVDANNSRASEINLK.LQHLELMEDIVDLAKK. | |
| Retrocopy | SKPISVLQDYCDAIKINIFWPLLFQHQNSSVISRLHPCVDANNSRASEINLKRLQHLELMEDIVDLAKKA |
| Parental | ANDSFLIGGLLRIGCKIENKILAMEEALNWIKYAGDVTILTKLGSIDNCWPMLSIFFTEYKYHITKIVME |
| ANDS.LIGGLLRIG.KIENKILAMEEALNWIKYAGDVTILTKLGSIDNCWPMLSIFFTEYKYHITKIVME | |
| Retrocopy | ANDSLLIGGLLRIGYKIENKILAMEEALNWIKYAGDVTILTKLGSIDNCWPMLSIFFTEYKYHITKIVME |
| Parental | DCNLLEELKTQSCMDCIEEGELMKMKGNEEFSKERFDIAIIYYTRAIEYRPENYLLYGNRALCFLRTGQF |
| DCNLLEELKTQSCMDCIE.GELMKMKGNEEFSKERFDIAIIYYTRAIEYRPEN.LLYGNRALCFLRTGQF | |
| Retrocopy | DCNLLEELKTQSCMDCIEQGELMKMKGNEEFSKERFDIAIIYYTRAIEYRPENHLLYGNRALCFLRTGQF |
| Parental | RNALGDGKRATILKNTWPKGHYRYCDALSMLGEYDWALQANIKAQKLCKNDPEGIKDLIQQHVKLQKQIE |
| RNALGDGKRATILKNTWPKGHYRYCDALSMLGEY.WALQANI.AQKLCKNDPEGIKDLIQQHVKLQKQIE | |
| Retrocopy | RNALGDGKRATILKNTWPKGHYRYCDALSMLGEYNWALQANITAQKLCKNDPEGIKDLIQQHVKLQKQIE |
| Parental | DLQGRTANKDPIKAFYENRAYTPRSLSAPIFTTSLNFVEKERDFRKINHEMANGGNQNLKVADEALKVDD |
| DLQGRTANK.PIKAFYENRAYTPRSLSAP.F.TSLNFVEKERDFRKINHEMANGGNQNLKV.DEAL.VDD | |
| Retrocopy | DLQGRTANKNPIKAFYENRAYTPRSLSAPVFSTSLNFVEKERDFRKINHEMANGGNQNLKVMDEALMVDD |
| Parental | CDCHPEFSPPSSQPPKHKGKQKSRNNESEKFSSSSPLTLPADLKNILEKQFSKSSRAAHQDFANIMKMLR |
| CDCHPEFSPPSSQPPKHKGKQKSRNNESEKFSSSS.LTLPADLKNILEKQFSKSSR.AHQDFANIM.MLR | |
| Retrocopy | CDCHPEFSPPSSQPPKHKGKQKSRNNESEKFSSSSQLTLPADLKNILEKQFSKSSRGAHQDFANIMTMLR |
| Parental | SLIQDGYMALLEQRCRSAAQAFTELLNGLDPQKIKQLNLAMINYVLVVYGLAISLLGIGQPEELSEAENQ |
| SLIQDGY.ALLEQRCRSAA.AFTELLNGLDPQKIKQLNLAMINYVLVVYGLAISLLGIGQPEELSEAENQ | |
| Retrocopy | SLIQDGYTALLEQRCRSAAHAFTELLNGLDPQKIKQLNLAMINYVLVVYGLAISLLGIGQPEELSEAENQ |
| Parental | FKRIIEHYPSEGLDCLAYCGIGKVYLKKNRFLEALNHFEKARTLIYRLPGVLTWPTSNVIIEESQPQKIK |
| FKRIIEHY..EGLDCLAYCGIGKVYL...RFLEALNHFEKARTLIYRLPGVLTWPTSNVIIEESQP.KIK | |
| Retrocopy | FKRIIEHYHNEGLDCLAYCGIGKVYLXXXRFLEALNHFEKARTLIYRLPGVLTWPTSNVIIEESQPEKIK |
| Parental | MLLEKFVEECKFPPVPDAICCYQKCHGYSKIQIYITDPDFKGFIRISCCQYCKIEFHMNCWKKLKTTTFN |
| MLLEKFVEECKFPPVPDAICCYQKC.GYSKIQIYITDPDFKGFIRISCCQYCKIEFHMNCWKKLKTTTFN | |
| Retrocopy | MLLEKFVEECKFPPVPDAICCYQKCCGYSKIQIYITDPDFKGFIRISCCQYCKIEFHMNCWKKLKTTTFN |
| Parental | DKIDKDFLQGICLTPDCEGVISKIIIFSSGGEVKCEFEHKVIKEKVPPRPILKQKCSSLEKLRLKEDKKL |
| DKIDKDFLQGICLTPDCEGVISKIIIFSSGG.VKCEFEHKVIKEKVPPRPILKQKCSSLEKLRLKED... | |
| Retrocopy | DKIDKDFLQGICLTPDCEGVISKIIIFSSGGQVKCEFEHKVIKEKVPPRPILKQKCSSLEKLRLKEDXXX |
| Parental | KRKIQKKEAKKLAQERMEEDLRESNPPKNEEQKETVDNVQRCQFLDDRILQCIKQYADKIKSGIQNTAML |
| ...........LAQERMEEDLRESNPPKNEEQKETVDNVQ.CQFLDDRILQCIKQYADKIKSGI.NTA.L | |
| Retrocopy | XXXXXXXXXXXLAQERMEEDLRESNPPKNEEQKETVDNVQHCQFLDDRILQCIKQYADKIKSGIWNTATL |
| Parental | LKELLSWKVLSTEDYTTCFSSRNFLNEAVDYVIRHLIQENNRVKTRIFLHVLSELKEVEPKLAAWIQKLN |
| LKELLSWKVLSTEDYTTCFSSRNFLNEAVDYVIRHLIQE.NRVKTRIFLHVLSELKEVEPKLAAWI.KLN | |
| Retrocopy | LKELLSWKVLSTEDYTTCFSSRNFLNEAVDYVIRHLIQEKNRVKTRIFLHVLSELKEVEPKLAAWIRKLN |
| Parental | SFGLDATGTFFSRYGASLKLLDFSIMTFLWNEKYGHKLDSIEGKQLDYFSEPASLKEARCLIWLLEEHRD |
| SFGLDATG.FFSRY.ASLK.LDFSIMTFLWNEKYGHKLDSIEGKQLDYF.EP.S.KEARCLIWLLEEHRD | |
| Retrocopy | SFGLDATGPFFSRYEASLKQLDFSIMTFLWNEKYGHKLDSIEGKQLDYFFEPTSSKEARCLIWLLEEHRD |
| Parental | KFPALHSALDEFFDIMDSRCTVLRKQDSGEAPFSSTKVKNKSKKKKPKDSKPMLVGSGTTSVTSNNEIIT |
| KFPALHSALDEFFDIMDSRCTVLRKQDSGEAPFSSTKVKNK.KKKKPKDSKPMLVGSGTTSVT.NNEIIT | |
| Retrocopy | KFPALHSALDEFFDIMDSRCTVLRKQDSGEAPFSSTKVKNKGKKKKPKDSKPMLVGSGTTSVTPNNEIIT |
| Parental | SSEDHSNRNSDSAGPFAVPDHLRQDVEEFEALYDQHSNEYVVRNKKLWDMNPKQKCSTLYDYFSQFLEEH |
| SSEDHSN.NSD.AGPFAVPDHLRQDVEEFEALYDQHSNEYVVRNKKLWDMNPKQKCSTLYDYFSQ.LEEH | |
| Retrocopy | SSEDHSNQNSDTAGPFAVPDHLRQDVEEFEALYDQHSNEYVVRNKKLWDMNPKQKCSTLYDYFSQLLEEH |
| Parental | GPLDMSNKMFSAEYEFFPEETRQILEKAGGLKPFLLGCPRFVVIDNCIALKKVASRLKKKRKKKNIKTKV |
| GPLDMS.KMFS.EYEFFPEETR.ILEKAGGLK.FLLGCPRFVVIDNCIALKKVASRL............. | |
| Retrocopy | GPLDMSHKMFSEEYEFFPEETRPILEKAGGLKSFLLGCPRFVVIDNCIALKKVASRLXXXXXXXXXXXXX |
| Parental | EEISKAGEYVRVKLQLNPAAREFKPDVKSKPVSDSSSA |
| E.ISKAGEY..VKL.LN.AAREFKPDVKSKPVS.SSSA | |
| Retrocopy | EDISKAGEYLQVKLPLNSAAREFKPDVKSKPVSGSSSA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 4 .61 RPM | 211 .78 RPM |
| bodymap2_adrenal | 3 .21 RPM | 139 .29 RPM |
| bodymap2_brain | 8 .79 RPM | 304 .25 RPM |
| bodymap2_breast | 6 .75 RPM | 285 .63 RPM |
| bodymap2_colon | 4 .76 RPM | 156 .27 RPM |
| bodymap2_heart | 2 .92 RPM | 70 .37 RPM |
| bodymap2_kidney | 3 .12 RPM | 126 .58 RPM |
| bodymap2_liver | 0 .46 RPM | 96 .34 RPM |
| bodymap2_lung | 2 .32 RPM | 98 .17 RPM |
| bodymap2_lymph_node | 2 .19 RPM | 144 .00 RPM |
| bodymap2_ovary | 10 .27 RPM | 292 .72 RPM |
| bodymap2_prostate | 3 .78 RPM | 216 .24 RPM |
| bodymap2_skeletal_muscle | 0 .38 RPM | 47 .09 RPM |
| bodymap2_testis | 6 .88 RPM | 245 .15 RPM |
| bodymap2_thyroid | 6 .55 RPM | 194 .43 RPM |
| bodymap2_white_blood_cells | 0 .39 RPM | 140 .49 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_4862 was not detected
15 EST(s) were mapped to retro_hsap_4862 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA470510 | 74963205 | 74963587 | 99.3 | 379 | 3 | 376 |
| AA470557 | 74963367 | 74963779 | 99.8 | 410 | 1 | 408 |
| AA502576 | 74963237 | 74963592 | 99.2 | 355 | 0 | 354 |
| AA554904 | 74963213 | 74963693 | 98.8 | 474 | 5 | 467 |
| AI678227 | 74963212 | 74963806 | 100 | 589 | 0 | 585 |
| AU117751 | 74964281 | 74964986 | 99.2 | 695 | 6 | 689 |
| AU127587 | 74964451 | 74965056 | 99.5 | 597 | 2 | 593 |
| AU144640 | 74964410 | 74964956 | 99.5 | 539 | 3 | 534 |
| BM701975 | 74963702 | 74964250 | 99.9 | 547 | 1 | 546 |
| BU729641 | 74963205 | 74963862 | 99.9 | 653 | 1 | 651 |
| BX495640 | 74963822 | 74964327 | 97.9 | 503 | 2 | 497 |
| DA200641 | 74964721 | 74965296 | 100 | 575 | 0 | 575 |
| DA494480 | 74964305 | 74964851 | 100 | 545 | 0 | 544 |
| DA509421 | 74964256 | 74964836 | 100 | 580 | 0 | 580 |
| DB355669 | 74963215 | 74963709 | 98.6 | 479 | 3 | 468 |
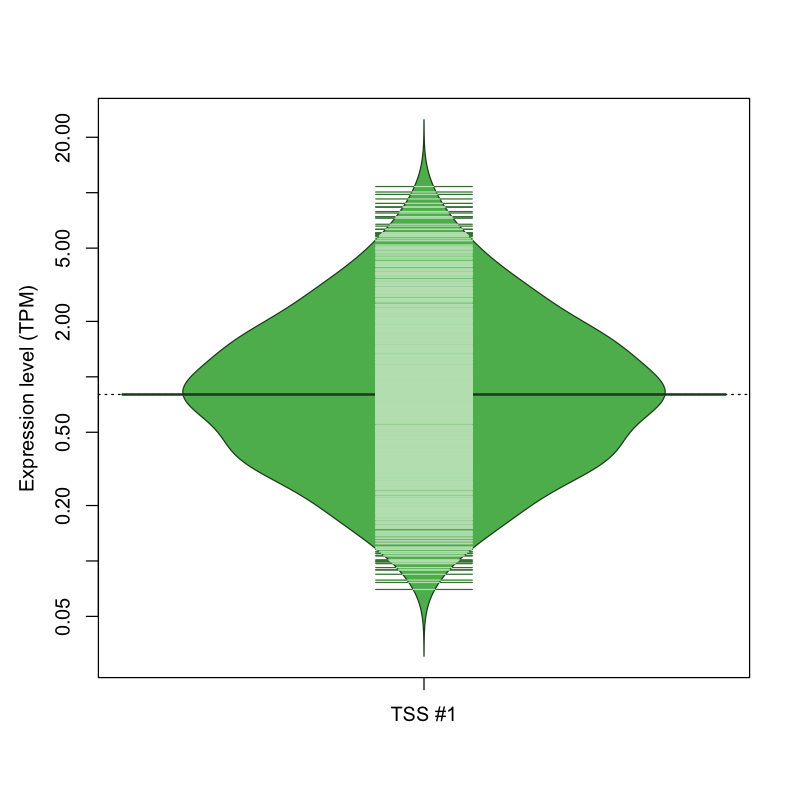
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_201219 | 866 libraries | 572 libraries | 358 libraries | 31 libraries | 2 libraries |
The graphical summary, for retro_hsap_4862 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_4862 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_4862 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_3032 |
Parental genes homology:
Parental genes homology involve 3 parental genes, and 4 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000182670 | 2 retrocopies |
retro_hsap_4862 , retro_hsap_886,
|
| Gorilla gorilla | ENSGGOG00000011116 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000004545 | 1 retrocopy |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .26 RPM |
| CEU_NA11843 | 0 .09 RPM |
| CEU_NA11930 | 0 .16 RPM |
| CEU_NA12004 | 0 .23 RPM |
| CEU_NA12400 | 0 .14 RPM |
| CEU_NA12751 | 0 .61 RPM |
| CEU_NA12760 | 0 .27 RPM |
| CEU_NA12827 | 0 .09 RPM |
| CEU_NA12872 | 0 .49 RPM |
| CEU_NA12873 | 0 .19 RPM |
| FIN_HG00183 | 0 .27 RPM |
| FIN_HG00277 | 0 .44 RPM |
| FIN_HG00315 | 0 .28 RPM |
| FIN_HG00321 | 0 .36 RPM |
| FIN_HG00328 | 0 .28 RPM |
| FIN_HG00338 | 0 .19 RPM |
| FIN_HG00349 | 0 .11 RPM |
| FIN_HG00375 | 0 .22 RPM |
| FIN_HG00377 | 0 .05 RPM |
| FIN_HG00378 | 0 .28 RPM |
| GBR_HG00099 | 0 .20 RPM |
| GBR_HG00111 | 0 .13 RPM |
| GBR_HG00114 | 0 .21 RPM |
| GBR_HG00119 | 0 .29 RPM |
| GBR_HG00131 | 0 .34 RPM |
| GBR_HG00133 | 0 .29 RPM |
| GBR_HG00134 | 0 .52 RPM |
| GBR_HG00137 | 0 .11 RPM |
| GBR_HG00142 | 0 .08 RPM |
| GBR_HG00143 | 0 .06 RPM |
| TSI_NA20512 | 0 .11 RPM |
| TSI_NA20513 | 0 .58 RPM |
| TSI_NA20518 | 0 .50 RPM |
| TSI_NA20532 | 0 .37 RPM |
| TSI_NA20538 | 0 .41 RPM |
| TSI_NA20756 | 0 .20 RPM |
| TSI_NA20765 | 0 .31 RPM |
| TSI_NA20771 | 0 .23 RPM |
| TSI_NA20786 | 0 .34 RPM |
| TSI_NA20798 | 0 .19 RPM |
| YRI_NA18870 | 0 .30 RPM |
| YRI_NA18907 | 0 .35 RPM |
| YRI_NA18916 | 0 .36 RPM |
| YRI_NA19093 | 0 .03 RPM |
| YRI_NA19099 | 0 .19 RPM |
| YRI_NA19114 | 0 .29 RPM |
| YRI_NA19118 | 0 .25 RPM |
| YRI_NA19213 | 0 .31 RPM |
| YRI_NA19214 | 0 .44 RPM |
| YRI_NA19223 | 0 .32 RPM |