RetrogeneDB ID: | retro_mmus_2037 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 2:60098402..60098699(-) | ||
| Located in intron of: | ENSMUSG00000026987 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000087181 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Mpc1 | ||
| Ensembl ID: | ENSMUSG00000023861 | ||
| Aliases: | Mpc1, 0610006G08Rik, 3830411I18Rik, Brp44l | ||
| Description: | mitochondrial pyruvate carrier 1 [Source:MGI Symbol;Acc:MGI:1915240] |

Retrocopy-Parental alignment summary:
>retro_mmus_2037
GCGCTAATGCGGAAAACGCGAGATTATTTTAAGACCAAGGAGTTTCGGGATTATATAACCAGCACCCACTTCTGGGGTCC
TGTGGCCAACTGGGGCCTTCCGCTGGCCGCCTTTAAGGATATGAAGGCGCCTCCCGACATCATCAGTGGCCGCATGACGA
CGGCCCTCATCTTCTACTCCATGGCTTTCATGCGCTTTGCCTACCGTGTCCAGCCTCGAAACTACCTGCTGCTGGCATGC
CATTTCTCCAACGTATTGGCGCAGAGCATCCAGGCGAGCCGTTACCTGAAGCACCAA
ORF - retro_mmus_2037 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 64.65 % |
| Parental protein coverage: | 90.83 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | ALVRKAADYVRSKDFRDYLMSTHFWGPVANWGLPIAAINDMKKSPEIISGRMTFALCCYSLTFMRFAYKV |
| AL.RK..DY...K.FRDY..STHFWGPVANWGLP.AA..DMK..P.IISGRMT.AL..YS..FMRFAY.V | |
| Retrocopy | ALMRKTRDYFKTKEFRDYITSTHFWGPVANWGLPLAAFKDMKAPPDIISGRMTTALIFYSMAFMRFAYRV |
| Parental | QPRNWLLFACHVTNEVAQLIQGGRLINYE |
| QPRN.LL.ACH..N..AQ.IQ..R..... | |
| Retrocopy | QPRNYLLLACHFSNVLAQSIQASRYLKHQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 13 .25 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 14 .63 RPM |
| SRP007412_heart | 0 .03 RPM | 32 .25 RPM |
| SRP007412_kidney | 0 .02 RPM | 22 .75 RPM |
| SRP007412_liver | 0 .05 RPM | 19 .01 RPM |
| SRP007412_testis | 5 .53 RPM | 2 .29 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_2037 was not detected
No EST(s) were mapped for retro_mmus_2037 retrocopy.
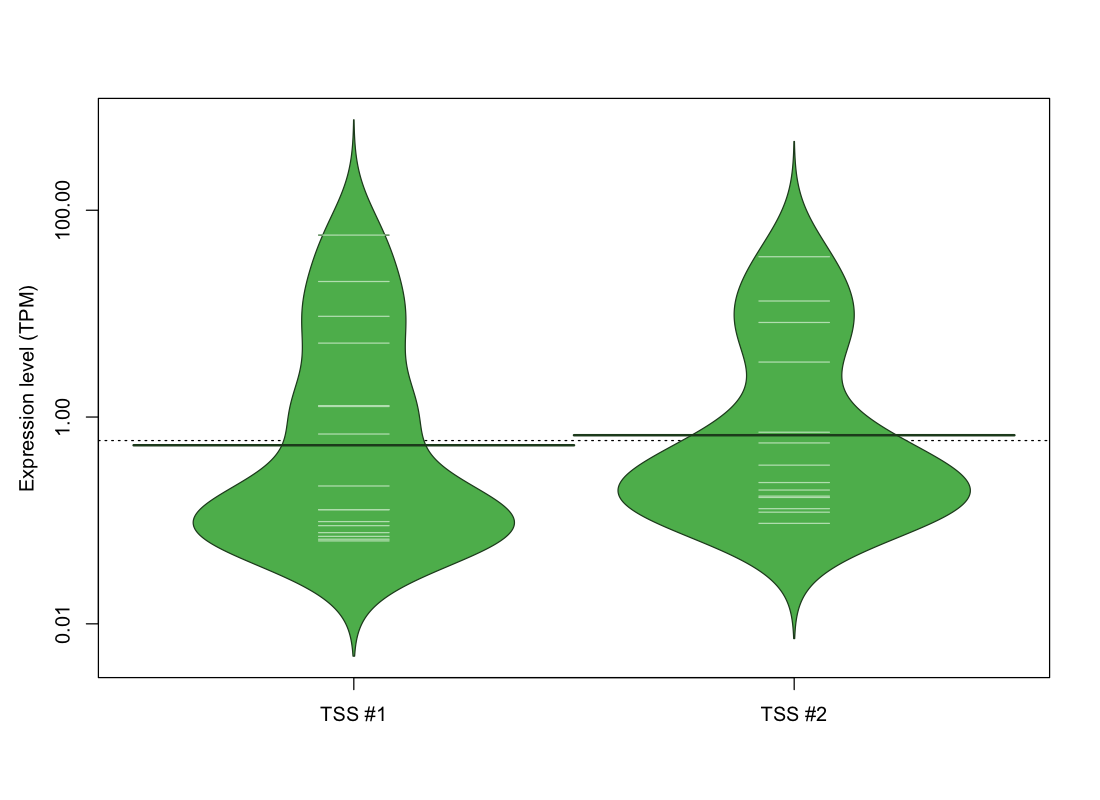
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_87825 | 1056 libraries | 10 libraries | 2 libraries | 2 libraries | 2 libraries |
| TSS #2 | TSS_87826 | 1058 libraries | 10 libraries | 1 library | 1 library | 2 libraries |
The graphical summary, for retro_mmus_2037 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2037 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2037 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 10 parental genes, and 15 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000027879 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000012552 | 2 retrocopies | |
| Dasypus novemcinctus | ENSDNOG00000001205 | 1 retrocopy | |
| Homo sapiens | ENSG00000060762 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000000583 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000023861 | 3 retrocopies |
retro_mmus_2037 , retro_mmus_3420, retro_mmus_979,
|
| Ornithorhynchus anatinus | ENSOANG00000004153 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000017166 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000012415 | 3 retrocopies | |
| Tursiops truncatus | ENSTTRG00000004456 | 1 retrocopy |