RetrogeneDB ID: | retro_mmus_241 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 7:60155608..60156103(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000047370 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ubald2 | ||
| Ensembl ID: | ENSMUSG00000050628 | ||
| Aliases: | Ubald2, 1110014K08Rik, D030012E24Rik, Fam100b | ||
| Description: | UBA-like domain containing 2 [Source:MGI Symbol;Acc:MGI:1914635] |

Retrocopy-Parental alignment summary:
>retro_mmus_241
ATGTCGGTGAACACGGACGAGCTGCGGCACCAGGTCATGATCAACCAGTTCGTGCTGGCCGCGGGCTGCGCGGCCGATCA
GGCGCAGCAGCTGCTGCAAGCGGCGCACTGGCAGTTCGAGACCGCCCTAAGCACGTTCTTTCAAGAAAGCAACATTCCCA
ACAGCCACCACCACCCGCAGATGATGTGTACCCCCAGCAACACCCCTGCTACACCACCCAACTTCCCTGATGCATTGGCC
ATGTTCTCCAAGCTTCGAACCTCTGAGGGCCTGCAGAGCAGCAGCAGCAGCCCCATGGCAGCCGTGGCCTGTTCCCCCCC
TGCAAACTTCAGCCCCTTCTGGGCCGCATCCCCACCCAATCACCAGGTGCCCTGGATCCCGCCCTCTTCCCCCAACACCT
TCCATCTCCACTGCCCACAGCCCACGTGGCCCCCCAGAGCATCGCAGGGGGGCGCCCCTCAGAAAGCCATGGCAGCCATG
GATGGCCAGAGATGA
ATGTCGGTGAACACGGACGAGCTGCGGCACCAGGTCATGATCAACCAGTTCGTGCTGGCCGCGGGCTGCGCGGCCGATCA
GGCGCAGCAGCTGCTGCAAGCGGCGCACTGGCAGTTCGAGACCGCCCTAAGCACGTTCTTTCAAGAAAGCAACATTCCCA
ACAGCCACCACCACCCGCAGATGATGTGTACCCCCAGCAACACCCCTGCTACACCACCCAACTTCCCTGATGCATTGGCC
ATGTTCTCCAAGCTTCGAACCTCTGAGGGCCTGCAGAGCAGCAGCAGCAGCCCCATGGCAGCCGTGGCCTGTTCCCCCCC
TGCAAACTTCAGCCCCTTCTGGGCCGCATCCCCACCCAATCACCAGGTGCCCTGGATCCCGCCCTCTTCCCCCAACACCT
TCCATCTCCACTGCCCACAGCCCACGTGGCCCCCCAGAGCATCGCAGGGGGGCGCCCCTCAGAAAGCCATGGCAGCCATG
GATGGCCAGAGATGA
ORF - retro_mmus_241 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 99.39 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSVNTDELRHQVMINQFVLAAGCAADQAQQLLQAAHWQFETALSTFFQESNIPNSHHHPQMMCTPSNTPA |
| MSVNTDELRHQVMINQFVLAAGCAADQAQQLLQAAHWQFETALSTFFQESNIPNSHHHPQMMCTPSNTPA | |
| Retrocopy | MSVNTDELRHQVMINQFVLAAGCAADQAQQLLQAAHWQFETALSTFFQESNIPNSHHHPQMMCTPSNTPA |
| Parental | TPPNFPDALAMFSKLRTSEGLQSSSSSPMAAVACSPPANFSPFWAASPPNHQVPWIPPSSPNTFHLHCPQ |
| TPPNFPDALAMFSKLRTSEGLQSSSSSPMAAVACSPPANFSPFWAASPPNHQVPWIPPSSPNTFHLHCPQ | |
| Retrocopy | TPPNFPDALAMFSKLRTSEGLQSSSSSPMAAVACSPPANFSPFWAASPPNHQVPWIPPSSPNTFHLHCPQ |
| Parental | PTWPPGASQGGAPQKAMAAMDGQR |
| PTWPP.ASQGGAPQKAMAAMDGQR | |
| Retrocopy | PTWPPRASQGGAPQKAMAAMDGQR |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 2 .22 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 0 .69 RPM |
| SRP007412_heart | 0 .03 RPM | 4 .64 RPM |
| SRP007412_kidney | 0 .12 RPM | 3 .70 RPM |
| SRP007412_liver | 0 .08 RPM | 1 .97 RPM |
| SRP007412_testis | 0 .09 RPM | 2 .97 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_241 was not detected
1 EST(s) were mapped to retro_mmus_241 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA061475 | 60155746 | 60155919 | 99.5 | 172 | 1 | 171 |
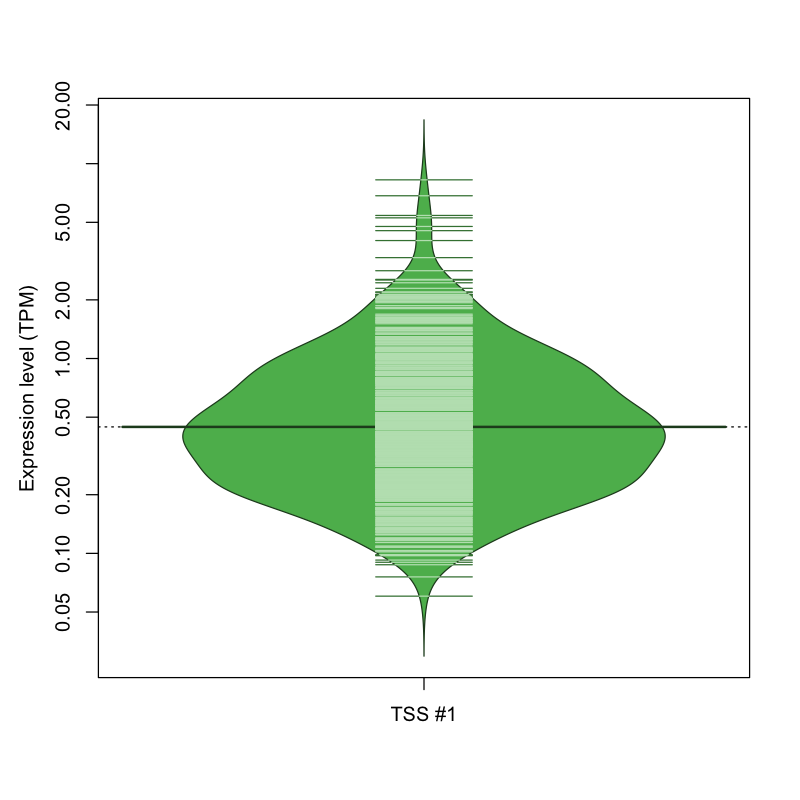
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_136443 | 459 libraries | 512 libraries | 97 libraries | 4 libraries | 0 libraries |
The graphical summary, for retro_mmus_241 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_241 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_241 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 3 parental genes, and 3 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Mus musculus | ENSMUSG00000039568 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000050628 | 1 retrocopy |
retro_mmus_241 ,
|
| Rattus norvegicus | ENSRNOG00000010029 | 1 retrocopy |