RetrogeneDB ID: | retro_mmus_245 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 15:81980559..81981012(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000061633 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Ndufb11 | ||
| Ensembl ID: | ENSMUSG00000031059 | ||
| Aliases: | Ndufb11, D5Bwg0566e, D5Bwg0577e, NP15.6, Np15 | ||
| Description: | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 [Source:MGI Symbol;Acc:MGI:1349919] |

Retrocopy-Parental alignment summary:
>retro_mmus_245
ATGGCGGCCTGGCGGTTAAGTTTGTGCGCTCGCAGCCTTTCCGCAATTCGACGGCTCCAGGCTGCCCCACTTTGCTGGAA
ATCCAGCTCCTCCAGGGCTGTGGTCGCCCGGTCCGCTGCGGCCCGAAAGGAGCAGGAAGAACCGACCGTGCAGTGGATGG
AGGACCCAGATCCCGAGGACGAAAACGTGTACTTGAAGAACCCACACTTCCATGGTTACGACGACGACCCGAAGGTGGAT
GCCTTGAATATGCACGCCGTCTTCTTCTTTGGTTTTTCCATCGCCCTGGTCCTTGGCACTACCTTTGTGGCTTATGTGCC
AAACTATAAGATGATCGAGTGGGCCCGCCGGGAAGCTGCGATGCTGGTGAAACACCGAGAGGCCAGTGGCCTCCCCATCA
TGGAGTCCAACTATTTTGACCCCACCAAGATCAACCTGCTGGAGGACGACTGA
ORF - retro_mmus_245 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 68.63 % |
| Parental protein coverage: | 100.0 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAARLLSLYGRCLSAAGAMRGLPAARVRWESS--RAVIAPSGVEKKRQREPTMQWQEDPEPEDENVYAKN |
| MAA..LSL..R.LS...A.R.L.AA...W.SS..RAV.A.S....K.Q.EPT.QW.EDP.PEDENVY.KN | |
| Retrocopy | MAAWRLSLCARSLS---AIRRLQAAPLCWKSSSSRAVVARSAAARKEQEEPTVQWMEDPDPEDENVYLKN |
| Parental | PDFHGYDSDPVVDVWNMRAVFFFGFSIVLVFGTTFVAYVPDYRMQEWARREAERLVKYREVNGLPIMESN |
| P.FHGYD.DP.VD..NM.AVFFFGFSI.LV.GTTFVAYVP.Y.M.EWARREA..LVK.RE..GLPIMESN | |
| Retrocopy | PHFHGYDDDPKVDALNMHAVFFFGFSIALVLGTTFVAYVPNYKMIEWARREAAMLVKHREASGLPIMESN |
| Parental | YFDPSKIQLPEDD |
| YFDP.KI.L.EDD | |
| Retrocopy | YFDPTKINLLEDD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 47 .12 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 55 .75 RPM |
| SRP007412_heart | 0 .00 RPM | 287 .39 RPM |
| SRP007412_kidney | 0 .00 RPM | 173 .27 RPM |
| SRP007412_liver | 0 .00 RPM | 121 .77 RPM |
| SRP007412_testis | 19 .94 RPM | 6 .63 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_245 was not detected
No EST(s) were mapped for retro_mmus_245 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_42858 | 1016 libraries | 17 libraries | 14 libraries | 15 libraries | 10 libraries |
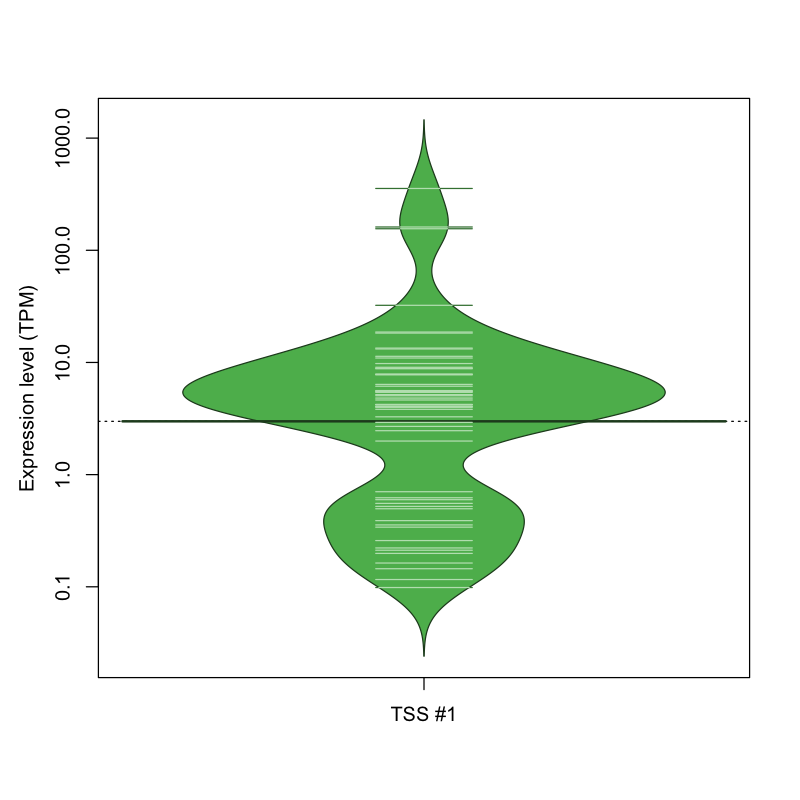
The graphical summary, for retro_mmus_245 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_245 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_245 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_151 |
Parental genes homology:
Parental genes homology involve 8 parental genes, and 10 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000147123 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000017422 | 2 retrocopies | |
| Monodelphis domestica | ENSMODG00000018491 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000031059 | 2 retrocopies |
retro_mmus_245 , retro_mmus_2602,
|
| Pongo abelii | ENSPPYG00000020284 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000021836 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000008329 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000026821 | 1 retrocopy |