RetrogeneDB ID: | retro_mmus_3665 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:94997393..94998704(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000087522 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Gmcl1 | ||
| Ensembl ID: | ENSMUSG00000001157 | ||
| Aliases: | Gmcl1, 2810049L19Rik, Gcl, mglc-1 | ||
| Description: | germ cell-less homolog 1 (Drosophila) [Source:MGI Symbol;Acc:MGI:1345156] |

Retrocopy-Parental alignment summary:
>retro_mmus_3665
AAAGTACCCCGTAGGACACTGTATGAATCACTGTTCCTGAGGGGTGAAAATAGCGATATAAAGATCTGTGCGTTTCAGGA
GGAGTGGCATTTGCACAAGTCCTATCTGTGCCGCTCTGGGTACTTTTCTAGCATGTTCTGTGGGGCCTGGCGAGAGTCAA
ATATGGATACGATAGAGATGCAGATGCCCGATGAAAATATTGATCGCGAGGCTTTCAATGATGTTCTAGGCTTCTTGTAC
AGTCGAAGAATTGTAATTCCTCCCCCTCGAGTCATTGCCGTCTTGGCCACAGCGAATATGCTGCAATTGGATGAGTTAAT
TCAGCAGTGTGAGGATATGATGAGAGCCTCAGTCAGTACCGAGACTGTGTGCAACTACTACTACTCTGCTGAGAATTACG
GGCTCCAGAGCATCAAATATGTTTGCTGCCAGTGGCTCTTGGACAACCTGATGACCCGAAAAAATGATGAACTATTGTTA
GGAATCAGTGTGGATCTCATGAAAGAGCTCATTGCCTCTTCAGATCTTTTGGTGGTTGGAGTGGAGATGGATGTGTATAT
TACACTGAAAACGTGGATGTACCTGCAGCTGGAAGCACGCACATGCTCAGGATCTGAAAGAGTATTACTGCCTGAAGAGT
TGAGCTTTGCAAAGTTCAGAAGGGAGTCAGATAACAGACCTTTTCTGGACAGTGATCAGGGGAGAGCCTTTGTGTCAGTG
TTCCAGCGACTAAGGCTTCCCTACATCATCAGTGACCTGCCTTCAGCACGCATTATTGAACAGGATAGACTGATCCCTGC
AGCATGGTTGACTCCAGTATACAAAGAGCAATGGCTAGCACTTCTTCAGGCAGAGCAGTCCAGGGAACTTGGGCGAATGG
ATATCAATGTGTCTGACATCCATGGAAACAGCATGAGGTGTGGAGGCCAACTTTTCACAGATGAGCAATGCAGCTGGACC
TGGTCTGGCTTCAACTTTGGCTGGGACTTGGTAGTGTGCTATAACAACAGGCGCATTATATTATGTCGCAGTGCAATGAA
TAAATCCTGTGGCCTTGGTGTCAGCTTACTCTGGCAGAGAAAAATTGCCTTCCGTCTACGAGTGATCTCATTGGACAGTA
CTGGAAAATCTGTTCTCAGAAGAGAGACTGACTATAATATTCTTTCTCTGAGAAAGAATCAACAACTAGAAGTGGTGAAC
CTCGGAAACCAAGATATAATTTTCCCCATATATGTGGCATGCAACTTCCTCTACCTTCCTGGAGAGAGTAGCGCTGGACC
AAGTGGGGAATCAAGCAGAAGTCCAATAAAC
AAAGTACCCCGTAGGACACTGTATGAATCACTGTTCCTGAGGGGTGAAAATAGCGATATAAAGATCTGTGCGTTTCAGGA
GGAGTGGCATTTGCACAAGTCCTATCTGTGCCGCTCTGGGTACTTTTCTAGCATGTTCTGTGGGGCCTGGCGAGAGTCAA
ATATGGATACGATAGAGATGCAGATGCCCGATGAAAATATTGATCGCGAGGCTTTCAATGATGTTCTAGGCTTCTTGTAC
AGTCGAAGAATTGTAATTCCTCCCCCTCGAGTCATTGCCGTCTTGGCCACAGCGAATATGCTGCAATTGGATGAGTTAAT
TCAGCAGTGTGAGGATATGATGAGAGCCTCAGTCAGTACCGAGACTGTGTGCAACTACTACTACTCTGCTGAGAATTACG
GGCTCCAGAGCATCAAATATGTTTGCTGCCAGTGGCTCTTGGACAACCTGATGACCCGAAAAAATGATGAACTATTGTTA
GGAATCAGTGTGGATCTCATGAAAGAGCTCATTGCCTCTTCAGATCTTTTGGTGGTTGGAGTGGAGATGGATGTGTATAT
TACACTGAAAACGTGGATGTACCTGCAGCTGGAAGCACGCACATGCTCAGGATCTGAAAGAGTATTACTGCCTGAAGAGT
TGAGCTTTGCAAAGTTCAGAAGGGAGTCAGATAACAGACCTTTTCTGGACAGTGATCAGGGGAGAGCCTTTGTGTCAGTG
TTCCAGCGACTAAGGCTTCCCTACATCATCAGTGACCTGCCTTCAGCACGCATTATTGAACAGGATAGACTGATCCCTGC
AGCATGGTTGACTCCAGTATACAAAGAGCAATGGCTAGCACTTCTTCAGGCAGAGCAGTCCAGGGAACTTGGGCGAATGG
ATATCAATGTGTCTGACATCCATGGAAACAGCATGAGGTGTGGAGGCCAACTTTTCACAGATGAGCAATGCAGCTGGACC
TGGTCTGGCTTCAACTTTGGCTGGGACTTGGTAGTGTGCTATAACAACAGGCGCATTATATTATGTCGCAGTGCAATGAA
TAAATCCTGTGGCCTTGGTGTCAGCTTACTCTGGCAGAGAAAAATTGCCTTCCGTCTACGAGTGATCTCATTGGACAGTA
CTGGAAAATCTGTTCTCAGAAGAGAGACTGACTATAATATTCTTTCTCTGAGAAAGAATCAACAACTAGAAGTGGTGAAC
CTCGGAAACCAAGATATAATTTTCCCCATATATGTGGCATGCAACTTCCTCTACCTTCCTGGAGAGAGTAGCGCTGGACC
AAGTGGGGAATCAAGCAGAAGTCCAATAAAC
ORF - retro_mmus_3665 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 51.95 % |
| Parental protein coverage: | 82.63 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | KSTSKYIYQTLFLNGENSDIKICALGEEWSLHKIYLCQSGYFSSMFSGSWKESSMNIIELEIPDQNIDIE |
| K......Y..LFL.GENSDIKICA..EEW.LHK.YLC.SGYFSSMF.G.W.ES.M..IE...PD.NID.E | |
| Retrocopy | KVPRRTLYESLFLRGENSDIKICAFQEEWHLHKSYLCRSGYFSSMFCGAWRESNMDTIEMQMPDENIDRE |
| Parental | ALQVAFGSLYRDDVLIKPSRVVAILAAACMLQLDGLIQQCGETMKETISVRTVCGYYTSAGTYGLDSVKK |
| A.....G.LY.....I.P.RV.A.LA.A.MLQLD.LIQQC...M....S..TVC.YY.SA..YGL.S.K. | |
| Retrocopy | AFNDVLGFLYSRRIVIPPPRVIAVLATANMLQLDELIQQCEDMMRASVSTETVCNYYYSAENYGLQSIKY |
| Parental | KCLEWLLNNLMTHQSVELFKELSINVMKQLIGSSNLFVMQVEMDVYTALKKWMFLQLVPSWNGSLKQLLT |
| .C..WLL.NLMT....EL....S...MK.LI.SS.L.V..VEMDVY..LK.WM.LQL...........L. | |
| Retrocopy | VCCQWLLDNLMTRKNDELLLGISVDLMKELIASSDLLVVGVEMDVYITLKTWMYLQLEARTCSGSERVLL |
| Parental | ETDVWFSKWKKDFEGTTFLETEQGKPFAPVFRHLRLQYIISDLASARIIEQDSLVPSEWLAAVYKQQWLA |
| .....F.K.........FL...QG..F..VF..LRL.YIISDL.SARIIEQD.L.P..WL..VYK.QWLA | |
| Retrocopy | PEELSFAKFRRESDNRPFLDSDQGRAFVSVFQRLRLPYIISDLPSARIIEQDRLIPAAWLTPVYKEQWLA |
| Parental | MLRAEQDSEVGPQEINKEELEGNSMRCGRKLAKDGEYCWRWTGFNFGFDLLVTYTNRYIIFKRNTLNQPC |
| .L.AEQ..E.G...IN.....GNSMRCG..L..D....W.W.GFNFG.DL.V.Y.NR.II..R...N..C | |
| Retrocopy | LLQAEQSRELGRMDINVSDIHGNSMRCGGQLFTDEQCSWTWSGFNFGWDLVVCYNNRRIILCRSAMNKSC |
| Parental | SGSVSLQPRRSIAFRLRLASFDSSGKLICSRATGYQILTLEKDQEQVVMNLDSRLLIFPLYICCNFLYIS |
| ...VSL...R.IAFRLR..S.DS.GK....R.T.Y.IL.L.K.Q...V.NL.....IFP.Y..CNFLY.. | |
| Retrocopy | GLGVSLLWQRKIAFRLRVISLDSTGKSVLRRETDYNILSLRKNQQLEVVNLGNQDIIFPIYVACNFLYLP |
| Parental | PEKRT----ESNRHPEN |
| .E.......ES.R.P.N | |
| Retrocopy | GESSAGPSGESSRSPIN |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 28 .04 RPM |
| SRP007412_cerebellum | 0 .04 RPM | 23 .40 RPM |
| SRP007412_heart | 0 .28 RPM | 5 .35 RPM |
| SRP007412_kidney | 0 .02 RPM | 20 .56 RPM |
| SRP007412_liver | 0 .00 RPM | 12 .32 RPM |
| SRP007412_testis | 0 .64 RPM | 112 .06 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_3665 was not detected
No EST(s) were mapped for retro_mmus_3665 retrocopy.
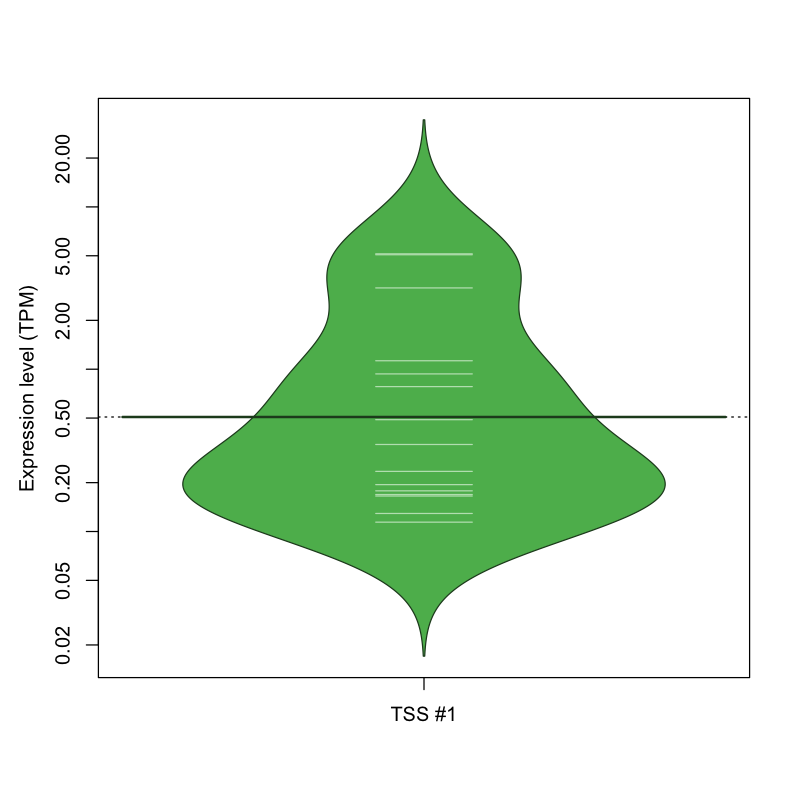
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_158570 | 1057 libraries | 11 libraries | 2 libraries | 2 libraries | 0 libraries |
The graphical summary, for retro_mmus_3665 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3665 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3665 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 19 parental genes, and 29 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000016116 | 1 retrocopy | |
| Choloepus hoffmanni | ENSCHOG00000012306 | 2 retrocopies | |
| Equus caballus | ENSECAG00000016813 | 2 retrocopies | |
| Homo sapiens | ENSG00000087338 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000002565 | 3 retrocopies | |
| Myotis lucifugus | ENSMLUG00000009183 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000000858 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000001157 | 1 retrocopy |
retro_mmus_3665 ,
|
| Nomascus leucogenys | ENSNLEG00000010132 | 1 retrocopy | |
| Oryctolagus cuniculus | ENSOCUG00000015808 | 2 retrocopies | |
| Otolemur garnettii | ENSOGAG00000008408 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000012334 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000012021 | 1 retrocopy | |
| Pteropus vampyrus | ENSPVAG00000001242 | 3 retrocopies | |
| Sorex araneus | ENSSARG00000004651 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000008335 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000000714 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000007493 | 3 retrocopies | |
| Vicugna pacos | ENSVPAG00000001756 | 1 retrocopy |