RetrogeneDB ID: | retro_mmus_3727 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:166482192..166483602(-) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000085880 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Fam154a | ||
| Ensembl ID: | ENSMUSG00000028492 | ||
| Aliases: | Fam154a, 4930500O09Rik | ||
| Description: | family with sequence similarity 154, member A [Source:MGI Symbol;Acc:MGI:1923061] |

Retrocopy-Parental alignment summary:
>retro_mmus_3727
AGGAAGTGCATCTGTGACCTTTGCTCCTGCGGCAGGCATCACTGCCCACATCTCCCTACCAAGATTTATGAGAAGACAGA
GAAACCATGTTTTTTCTCCGAATACTCGGAGAAGTATTCTACCTATCATTCTTATATGCCCCAAGAGTCCTTCAAACCCA
AGGTGGAGTACCAGAAAGTAGCTATACCAATGGAAGGACTGTCCACAACAAAGAGAGACTTTGGGGCTTACAACATAGTA
CCGGTGAAACACCATCTTCCCGAGAAGGTCACTCCAATCCAAGATGAAATGGATTTCCTCACAACGTATAATCAACACTA
CAACTATTGCCCTGCCACTCGAGCGAGCCCCATCAAACCTCGAGACAACAGGCATCAATGTAATGACAAAATGGAGTATG
AGCCTACGTACAAAGCTGATTATTTACCTTGGAACCAACAAAAAAGATCTTTGATTCATCCACCACAATCATACCGTCCT
CCGTCCTGTAGGTTTGATCATAGAACCACACACCAGGACGATTATCCCATGAAAAGTCCTGTGGATACTGTGAATTACAA
ACCTCCACCTGGGCCCAAGCTCTCCAACCTCCCCCTGGAGAATATGACAAGCTACAGATCGAGCTATGTGGCCCATCCTG
TGGAGAAGCGGTGTGTGTATGAGGGAGAGAAATATAAACCATGTGAAATTCCTTTTGATGGCCTTACCACCCACAGAGAT
TCCTACAAGGGTTTGATGGGGGAACCTGCCAAGAGCTGGAAACCTGTACCAAATCACTCTGGGCTAGGTATACCTTTTGC
AACCGACACTGAATTTCGAGAAAATTTTCAAGCTTGGCCAACACCTAAGATTGTCCCCAAAGAGCCCATTCCCTACATCC
CTCCTGAAGGAAAGATGGATCTTCTGACAACTGTGCAAGCCAACTATAAATGCCCCAATGGCGCTCCTGCTCAGTCCTGC
CGGCCAGTCATCCACTTGAAGAAGAGTGAGCGATTTGAAAGCTCTACCACCAACAGAGAGGATTATAAACACTGGGACAT
CATCCCTAGAGAGCCAATCAAACCTGCTCCCCAGCTGAAATTCCCTGATGAGCCTATGGATTACATGACCACCAATCGTG
CCCACTACGTGCCTCATGCACCAGTCAATACCAAAAGCTGCAAGCCAACCTGGTCCGGCCCTCGTGTAAACATCCCGTTA
GAAGGTCAGACCACCTACTCTTCTAATTTTACTCCTAAGGAAATTCAAAGGTGCCCAGCTTCATTCTCTGAACCCCCTGG
CTACATCTTTGAAGAGGTGGATGCTGTAGGGCACAGGATGTACAGGCCGGCTTCCAGAGCAGCTTCTCATGAGAGCAACC
ATCTTGGCTTTGGTGATGTGTAAAGAATTGGCACTCCCAGCCTGTCATTG
ORF - retro_mmus_3727 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 54.99 % |
| Parental protein coverage: | 98.95 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | RTCICNLCSCGRHRCPHLPTKIYDKTEKTRLLSEYTENYPIYQSYLPRNSFKPEWCYRKPSAPMEGLTTC |
| R.CIC.LCSCGRH.CPHLPTKIY.KTEK....SEY.E.Y..Y.SY.P..SFKP...Y.K...PMEGL.T. | |
| Retrocopy | RKCICDLCSCGRHHCPHLPTKIYEKTEKPCFFSEYSEKYSTYHSYMPQESFKPKVEYQKVAIPMEGLSTT |
| Parental | RRDFGLHKMLPVTFHQPDTYVPSQETMDLLTTYKHDFNYLPTCPVGLIKPRDSKFPNGDKIMQYLPTYKV |
| .RDFG.....PV..H.P....P.Q..MD.LTTY....NY.P......IKPRD......DK.M.Y.PTYK. | |
| Retrocopy | KRDFGAYNIVPVKHHLPEKVTPIQDEMDFLTTYNQHYNYCPATRASPIKPRDNRHQCNDK-MEYEPTYKA |
| Parental | DYLPWNQPRRELLRPPHKYRPESSKFENRTTHQDDYTMKGLVTTVSCKPPSKSKIYNIPLEDLTNYKMSY |
| DYLPWNQ..R.L..PP..YRP.S..F..RTTHQDDY.MK..V.TV..KPP...K..N.PLE..T.Y..SY | |
| Retrocopy | DYLPWNQQKRSLIHPPQSYRPPSCRFDHRTTHQDDYPMKSPVDTVNYKPPPGPKLSNLPLENMTSYRSSY |
| Parental | VPHPVEKRFVKEAEKYIPCDIPFENLTTHKESYRGLLGEPAKSTKPPAKFPVHDAPFTNTTEVQEKFQAW |
| V.HPVEKR.V.E.EKY.PC.IPF..LTTH..SY.GL.GEPAKS.KP.........PF...TE..E.FQAW | |
| Retrocopy | VAHPVEKRCVYEGEKYKPCEIPFDGLTTHRDSYKGLMGEPAKSWKPVPNHSGLGIPFATDTEFRENFQAW |
| Parental | PTPQMVPKAPVVYVPPEEKMDLLTTVQTHYKYLKGSPAITCRPVPSIKKSKCFESSTTTKDDFKQWAAVN |
| PTP..VPK.P..Y.PPE.KMDLLTTVQ..YK...G.PA..CRPV...KKS..FESSTT...D.K.W.... | |
| Retrocopy | PTPKIVPKEPIPYIPPEGKMDLLTTVQANYKCPNGAPAQSCRPVIHLKKSERFESSTTNREDYKHWDIIP |
| Parental | TKPIRPTLHLSLPAEPLDCQTTTKTCFVAHPPIITENYKPPWVGLRRNIPVEGQTTYSINFTPKDLGKCP |
| ..PI.P...L..P.EP.D..TT.....V.H.P..T...KP.W.G.R.NIP.EGQTTYS.NFTPK....CP | |
| Retrocopy | REPIKPAPQLKFPDEPMDYMTTNRAHYVPHAPVNTKSCKPTWSGPRVNIPLEGQTTYSSNFTPKEIQRCP |
| Parental | ASYMEPPGYIFEEVDSMGHKIYRSISQTGSRQNSSISICDTEKPGQKELEV |
| AS..EPPGYIFEEVD..GH..YR..S...S.........D....G...L.. | |
| Retrocopy | ASFSEPPGYIFEEVDAVGHRMYRPASRAASHESNHLGFGDV*RIGTPSLSL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 0 .12 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 0 .00 RPM |
| SRP007412_heart | 0 .00 RPM | 0 .00 RPM |
| SRP007412_kidney | 0 .02 RPM | 0 .04 RPM |
| SRP007412_liver | 0 .00 RPM | 0 .03 RPM |
| SRP007412_testis | 12 .58 RPM | 63 .21 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_3727 was not detected
No EST(s) were mapped for retro_mmus_3727 retrocopy.
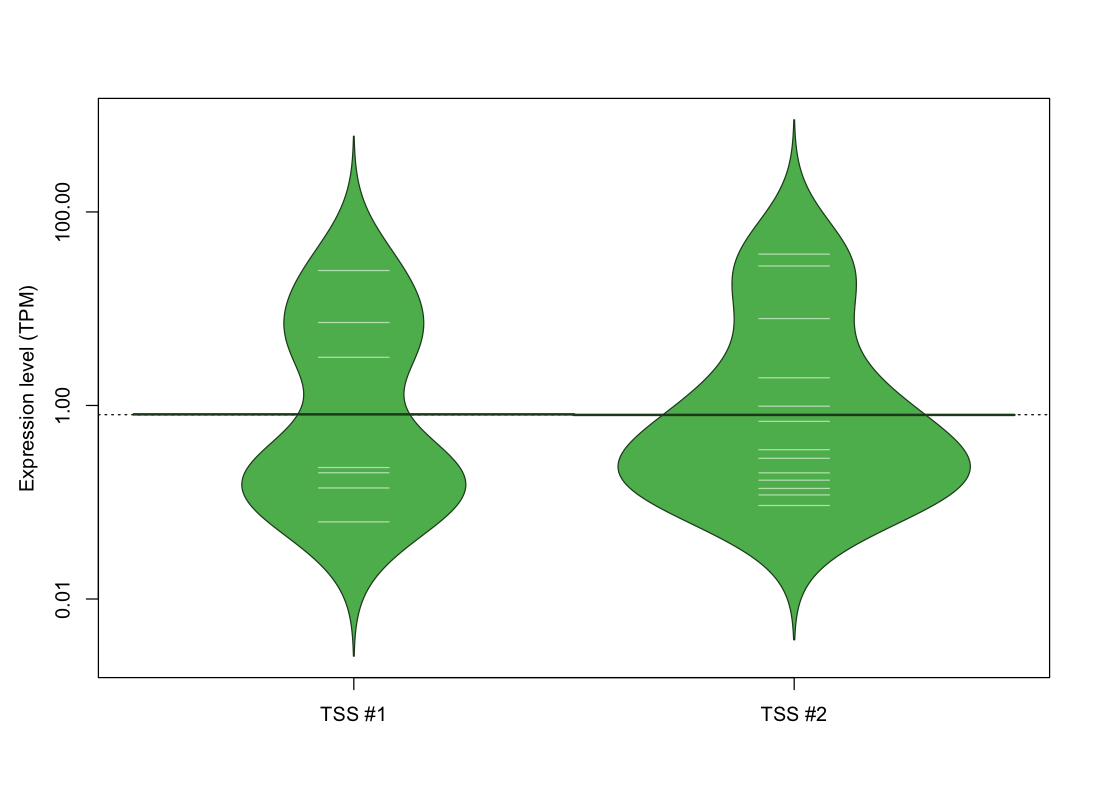
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_156360 | 1065 libraries | 4 libraries | 1 library | 1 library | 1 library |
| TSS #2 | TSS_156361 | 1059 libraries | 9 libraries | 1 library | 1 library | 2 libraries |
The graphical summary, for retro_mmus_3727 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_3727 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3727 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_20 |
Parental genes homology:
Parental genes homology involve 2 parental genes, and 2 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Mus musculus | ENSMUSG00000028492 | 1 retrocopy |
retro_mmus_3727 ,
|
| Rattus norvegicus | ENSRNOG00000048197 | 1 retrocopy |