RetrogeneDB ID: | retro_mmus_1668 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 17:79933511..79934572(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Eif3h | ||
| Ensembl ID: | ENSMUSG00000022312 | ||
| Aliases: | Eif3h, 1110008A16Rik, 40kD, 9430017H16Rik, EIF3-P40, EIF3-gamma, Eif3s3 | ||
| Description: | eukaryotic translation initiation factor 3, subunit H [Source:MGI Symbol;Acc:MGI:1915385] |

Retrocopy-Parental alignment summary:
>retro_mmus_1668
ATGGCGTCGCGCAAGGAAGGCACCGGCTCCACTGCCACCTCCTCCAGTTCTGCTGGCGGCACGGTGGGGAAGGGCAAAGG
CAAGGGCGGCTCTGGAGATTCGGTGGTGAAGCAGGTGCAGATCGACGGCCTGGTAGTATTAAAGATAATCAAACATTATC
AAGAAGAAGGACAAGGAACTGAGGTTGTTCAGGGAGTGCTCCTGGGTCTGGTTGTAGAAGATCGACTAGAGATGACCAAC
TGCTTCCCGTTCCCCCCAGCACACGGAGGATGATGCTGACTTTGATGAGGTACAGTATCAGATGGAGATGATGCGCAGCC
TTCGCCATGTCAACATTGATCACCTCCACGTGGGCTGGTATCAGTCCACATATTACGGCTCCTTCGTTACCCGGGCGCTT
CTGGATTCTCAGTTCAGCTACCAGCACGCCATTGAAGAGTCTGTCGTCCTCATTTATGATCCCATAAAAACTGCCCAAGG
ATCTCTCTCTCGCTGAAGGCATACATGACTGACTCCTAAACGGATGGAAGTTTGTAAAGAGAAGGACTTTTCCCCTGAAG
CACTGAAAAAGGCAAGCATCACCTTTGAGCACATGTTTGAAGAAGTGCCGATTGTGATTAAAAACTCACATCTGATCAGT
GTCCTTATGTGGGAGCTTGAGAAGAAGTCAGCTGTGGCGGATAAGCACGAATTGCTCAGTCTTGCTAGCAGCAATCATCT
GGGGGAAGAATCTCCAGCTGCTGATGGACCCGGTGGATGAAATGAGCCAGGACATAATCAAATACAACACAGACGTGCGG
AACACCAGTAAACAGCAGCAGCAGAAACATCAGAATCAGCAGCGCCGCCAACAGGAGAATATGCAGCGTCGGAGTCGAGG
CGAGCCCCCACTCCCTGAGGAGGACCTCTCTAAACTCTTCAAGCCCCACCAGGCCCCTGCCAGGATGGATTCACTGCTCA
TTGCAGGCCAGATGAACACTTACTGCCAGAACATCAAGGAGTTCACTGCCCAAAACTTAGGCAAACTCTTCATGGCCCAG
GCTCTTCAAGAATACAATAAT
ATGGCGTCGCGCAAGGAAGGCACCGGCTCCACTGCCACCTCCTCCAGTTCTGCTGGCGGCACGGTGGGGAAGGGCAAAGG
CAAGGGCGGCTCTGGAGATTCGGTGGTGAAGCAGGTGCAGATCGACGGCCTGGTAGTATTAAAGATAATCAAACATTATC
AAGAAGAAGGACAAGGAACTGAGGTTGTTCAGGGAGTGCTCCTGGGTCTGGTTGTAGAAGATCGACTAGAGATGACCAAC
TGCTTCCCGTTCCCCCCAGCACACGGAGGATGATGCTGACTTTGATGAGGTACAGTATCAGATGGAGATGATGCGCAGCC
TTCGCCATGTCAACATTGATCACCTCCACGTGGGCTGGTATCAGTCCACATATTACGGCTCCTTCGTTACCCGGGCGCTT
CTGGATTCTCAGTTCAGCTACCAGCACGCCATTGAAGAGTCTGTCGTCCTCATTTATGATCCCATAAAAACTGCCCAAGG
ATCTCTCTCTCGCTGAAGGCATACATGACTGACTCCTAAACGGATGGAAGTTTGTAAAGAGAAGGACTTTTCCCCTGAAG
CACTGAAAAAGGCAAGCATCACCTTTGAGCACATGTTTGAAGAAGTGCCGATTGTGATTAAAAACTCACATCTGATCAGT
GTCCTTATGTGGGAGCTTGAGAAGAAGTCAGCTGTGGCGGATAAGCACGAATTGCTCAGTCTTGCTAGCAGCAATCATCT
GGGGGAAGAATCTCCAGCTGCTGATGGACCCGGTGGATGAAATGAGCCAGGACATAATCAAATACAACACAGACGTGCGG
AACACCAGTAAACAGCAGCAGCAGAAACATCAGAATCAGCAGCGCCGCCAACAGGAGAATATGCAGCGTCGGAGTCGAGG
CGAGCCCCCACTCCCTGAGGAGGACCTCTCTAAACTCTTCAAGCCCCACCAGGCCCCTGCCAGGATGGATTCACTGCTCA
TTGCAGGCCAGATGAACACTTACTGCCAGAACATCAAGGAGTTCACTGCCCAAAACTTAGGCAAACTCTTCATGGCCCAG
GCTCTTCAAGAATACAATAAT
ORF - retro_mmus_1668 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 94.08 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 2 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MASRKEGTGSTATSSGSAGGAVGKGKGKGGSGDSAVKQVQIDGLVVLKIIKHYQEEGQGTEVVQGVLLGL |
| MASRKEGTGSTATSS.SAGG.VGKGKGKGGSGDS.VKQVQIDGLVVLKIIKHYQEEGQGTEVVQGVLLGL | |
| Retrocopy | MASRKEGTGSTATSSSSAGGTVGKGKGKGGSGDSVVKQVQIDGLVVLKIIKHYQEEGQGTEVVQGVLLGL |
| Parental | VVEDRLEITNCFPF-PQHTEDDADFDEVQYQMEMMRSLRHVNIDHLHVGWYQSTYYGSFVTRALLDSQFS |
| VVEDRLE.TNCFPF.PQHTEDDADFDEVQYQMEMMRSLRHVNIDHLHVGWYQSTYYGSFVTRALLDSQFS | |
| Retrocopy | VVEDRLEMTNCFPF>PQHTEDDADFDEVQYQMEMMRSLRHVNIDHLHVGWYQSTYYGSFVTRALLDSQFS |
| Parental | YQHAIEESVVLIYDPIKTAQGSLSLKAYR-LTPKLMEVCKEKDFSPEALKKASITFEHMFEEVPIVIKNS |
| YQHAIEESVVLIYDPIKTAQGSLS......LTPK.MEVCKEKDFSPEALKKASITFEHMFEEVPIVIKNS | |
| Retrocopy | YQHAIEESVVLIYDPIKTAQGSLSR*RHT*LTPKRMEVCKEKDFSPEALKKASITFEHMFEEVPIVIKNS |
| Parental | HLINVLMWELEKKSAVADKHELLSLASSNHL-GKSLQLLMDRVDEMSQDIIKYNTYMRNTSKQQQQKHQY |
| HLI.VLMWELEKKSAVADKHELLSLASSNHL.GK.LQLLMD.VDEMSQDIIKYNT..RNTSKQQQQKHQ. | |
| Retrocopy | HLISVLMWELEKKSAVADKHELLSLASSNHL>GKNLQLLMDPVDEMSQDIIKYNTDVRNTSKQQQQKHQN |
| Parental | QQRRQQENMQRQSRGEPPLPEEDLSKLFKPHQAPARMDSLLIAGQINTYCQNIKEFTAQNLGKLFMAQAL |
| QQRRQQENMQR.SRGEPPLPEEDLSKLFKPHQAPARMDSLLIAGQ.NTYCQNIKEFTAQNLGKLFMAQAL | |
| Retrocopy | QQRRQQENMQRRSRGEPPLPEEDLSKLFKPHQAPARMDSLLIAGQMNTYCQNIKEFTAQNLGKLFMAQAL |
| Parental | QEYNN |
| QEYNN | |
| Retrocopy | QEYNN |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 70 .11 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 95 .44 RPM |
| SRP007412_heart | 0 .19 RPM | 117 .92 RPM |
| SRP007412_kidney | 0 .17 RPM | 105 .30 RPM |
| SRP007412_liver | 0 .11 RPM | 95 .31 RPM |
| SRP007412_testis | 0 .73 RPM | 123 .68 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_1668 was not detected
No EST(s) were mapped for retro_mmus_1668 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_56876 | 412 libraries | 50 libraries | 304 libraries | 250 libraries | 56 libraries |
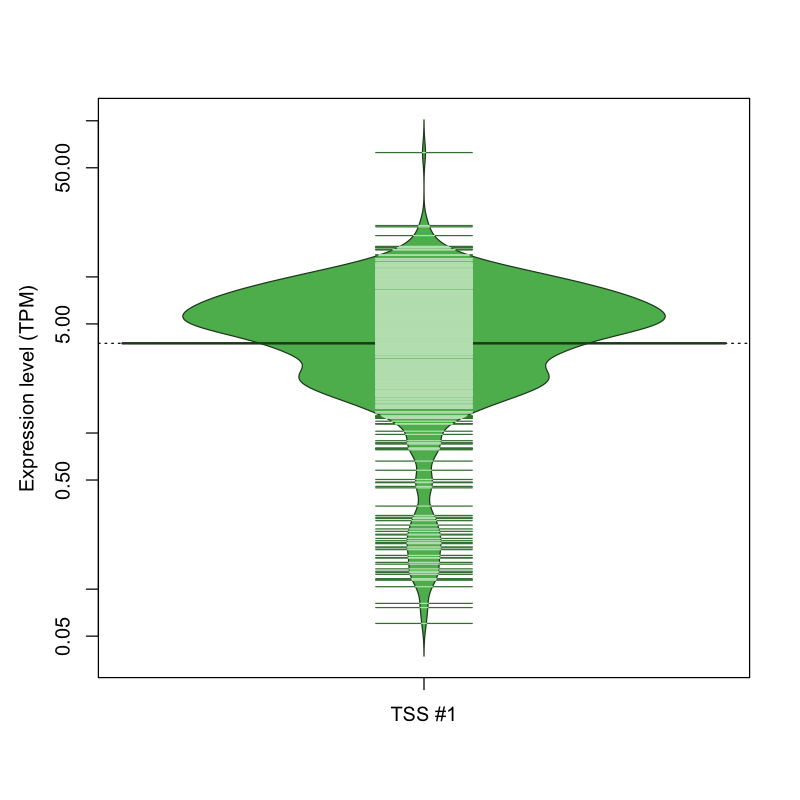
The graphical summary, for retro_mmus_1668 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1668 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1668 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 14 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Canis familiaris | ENSCAFG00000000816 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000005910 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000000324 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000015663 | 4 retrocopies | |
| Monodelphis domestica | ENSMODG00000004154 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000022312 | 1 retrocopy |
retro_mmus_1668 ,
|
| Oryctolagus cuniculus | ENSOCUG00000007534 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000008905 | 1 retrocopy | |
| Sorex araneus | ENSSARG00000009067 | 1 retrocopy | |
| Sarcophilus harrisii | ENSSHAG00000014977 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000001403 | 1 retrocopy |