RetrogeneDB ID: | retro_mmus_207 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 6:50608636..50610709(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000092012 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Crnkl1 | ||
| Ensembl ID: | ENSMUSG00000001767 | ||
| Aliases: | Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn | ||
| Description: | Crn, crooked neck-like 1 (Drosophila) [Source:MGI Symbol;Acc:MGI:1914127] |

Retrocopy-Parental alignment summary:
>retro_mmus_207
ATGGCGTCATCCACCGTGGCCAAGAAAGAGCAGCGCCTTCCTAAAGTGGCCCAGGTCAAAAACAAAGCTGCGGCCGAGGT
GCAGATAACTGCAGAGCAGCTCTTAAGGGAAGCTAAAGAGAGAGAACTCGAGCTCCTCGCGCCTCCACCTCAGCAGAAGA
TCACAGACGCAGAAGAATTCAATGATTACAAACTCCGGAGAAGAAAGGCCTTTGAAGAAAACCTCAGACAGAAGAGGACT
GTGATTAGCAACTGGATCAAATACGCACAATGGGAGGAAAGCCTGAAGGAGCTCGATAGGGCTCGATCCATATACGAGCG
TGCTTTAGATGTGGACTACCGCAATCTTACCCTCTGGCTGAAATACGCAGAGATGGAGATGAAGAACCGCCAGGTCAACC
ATGCCCGAAACATCTGGGACCGTGCCATCACAACTCTGCCCCGAGTCAAGCAGTTGTGGTACAAGTACACGTACATGGAG
GAGCTGTTGGGCAACGTCGCTGGTGCTCGTCAGGTGTTTGAGCGCTGGATGGAATGGCAGCCTGAGGAGCAGGCCTGGCA
CTCGTACATCAACTTGGAGCTGAGGTACAAAGAGGTGGAGCGGGCCCGCACCATTTATGAACGCTTTGTGCTCGTGCACC
CTGCTGTGAAGAACTGGATCAAGTATGCTCGGTTTGAAGAGCGGCACGCTTCTGTTGCTCATGCGCGGAAAGTCTACGAG
AGGGCCGTGGAGTTTTTCGGGGAGGAGCATATGGACCAACACCTCTATGTGGCCTTTGCAAAATTCGAGGAGAATCAGAA
AGAACTGGAAAGGGTGCGAGTTATCTACAGATATGCCCTGGATAGGATTTCCAAACAGGAGGCCCAAGAACTCTTGACAA
ACTACACCATCTTTGAGAAGAAGTTTGGTGAGCGGAGGGATGTAGAAGAGAGCATCGTGAGAAAACGCCGATTCCACTAC
GAAGAAGAAGTGAAGGCTAATCCGCACAATTACGACGCGTGGTTTGATTACTTACGCTTGGTGGAAAGTGAGGCAGAAGC
CGACACCGTGCGGGAAGTCTATGAGAGAGCCATCGCCAGTGTGCCACCGGTGCTGGAGAAGAGGCACTGGAAGCGCTACA
TCTACCTCTGGGTTAACTATGCGCTCTATGAAGAGCTGGAGGCCAAGGATCCCGAGAGGACGAGACGCGTCTATCAAGCG
TCTCTGCAGCTCATTCCTCACACAAAGTTCACCTTTGCCAAAATGTGGTTGTATTATGCCCAGTTTGAAATACGGCAGAA
GAATCTGCCATTGGCCAGAAGAGCTTTGGGCACTTCCCTAGGCAAATGTCCAAAGAACAAGTTGTTCAAAGGCTACATAG
CCCTGGAGCTGCAGCTTCGAGAGTTTGACAGATGCAGGAAGCTTTATGAGAAGTTTCTGGAATTCGGACCTGAAAATTGT
ACCTCGTGGATAAAGTTTGCAGAGTTAGAGACAATCCTTGGTGATGTTGAGAGAGCTCGGGCAATCTATGAGTTAGCCAT
CAGCCAGCCGCGCTTGGATATGCCAGAGCTACTTTGGAAATCCTATATTGATTTTGAGATGGAGCAGGAAGAGCCTGAGA
GAACACGGAACCTTTACCGACGGTTGCTTCAGAGAACACAGCACGTCAAGGTGTGGATCAGTTTGGCTCAGTTTGAGTTC
TCTTCTGGGAAAGAAGGAAGTGTGGCGAAATGCAGACAGATTTATCAAGAAGCCAACAGATCCATGAGACACTGCCAGGA
CAAAGAAGAGAGGCTTATGCTGTTGGAATCCTGGAGAAGTTTTGAAGGGGAGTTTGGAACCGTAGCAGACAAGGAGAGAG
TAGACAGACTCATGCCAGACGAGGTCAAGAAGAGAAGAAAGGTCCAGGCCGACGATGGTTCTGATGCCGGATGGCAAGAA
TACCGGGACTACATCTTTCCAGAGGATGCGGCCCAGCAGCCCAACCTCAAGCTTCTGGCCGTGGCCAAGCTGTGGAAGAA
ACAGCAACAGGAAAGAGAAGCTGCCGGACAGGATCCAGATAAGGACAGGAATGGGAGTGCACTTGCTGGTTAC
ATGGCGTCATCCACCGTGGCCAAGAAAGAGCAGCGCCTTCCTAAAGTGGCCCAGGTCAAAAACAAAGCTGCGGCCGAGGT
GCAGATAACTGCAGAGCAGCTCTTAAGGGAAGCTAAAGAGAGAGAACTCGAGCTCCTCGCGCCTCCACCTCAGCAGAAGA
TCACAGACGCAGAAGAATTCAATGATTACAAACTCCGGAGAAGAAAGGCCTTTGAAGAAAACCTCAGACAGAAGAGGACT
GTGATTAGCAACTGGATCAAATACGCACAATGGGAGGAAAGCCTGAAGGAGCTCGATAGGGCTCGATCCATATACGAGCG
TGCTTTAGATGTGGACTACCGCAATCTTACCCTCTGGCTGAAATACGCAGAGATGGAGATGAAGAACCGCCAGGTCAACC
ATGCCCGAAACATCTGGGACCGTGCCATCACAACTCTGCCCCGAGTCAAGCAGTTGTGGTACAAGTACACGTACATGGAG
GAGCTGTTGGGCAACGTCGCTGGTGCTCGTCAGGTGTTTGAGCGCTGGATGGAATGGCAGCCTGAGGAGCAGGCCTGGCA
CTCGTACATCAACTTGGAGCTGAGGTACAAAGAGGTGGAGCGGGCCCGCACCATTTATGAACGCTTTGTGCTCGTGCACC
CTGCTGTGAAGAACTGGATCAAGTATGCTCGGTTTGAAGAGCGGCACGCTTCTGTTGCTCATGCGCGGAAAGTCTACGAG
AGGGCCGTGGAGTTTTTCGGGGAGGAGCATATGGACCAACACCTCTATGTGGCCTTTGCAAAATTCGAGGAGAATCAGAA
AGAACTGGAAAGGGTGCGAGTTATCTACAGATATGCCCTGGATAGGATTTCCAAACAGGAGGCCCAAGAACTCTTGACAA
ACTACACCATCTTTGAGAAGAAGTTTGGTGAGCGGAGGGATGTAGAAGAGAGCATCGTGAGAAAACGCCGATTCCACTAC
GAAGAAGAAGTGAAGGCTAATCCGCACAATTACGACGCGTGGTTTGATTACTTACGCTTGGTGGAAAGTGAGGCAGAAGC
CGACACCGTGCGGGAAGTCTATGAGAGAGCCATCGCCAGTGTGCCACCGGTGCTGGAGAAGAGGCACTGGAAGCGCTACA
TCTACCTCTGGGTTAACTATGCGCTCTATGAAGAGCTGGAGGCCAAGGATCCCGAGAGGACGAGACGCGTCTATCAAGCG
TCTCTGCAGCTCATTCCTCACACAAAGTTCACCTTTGCCAAAATGTGGTTGTATTATGCCCAGTTTGAAATACGGCAGAA
GAATCTGCCATTGGCCAGAAGAGCTTTGGGCACTTCCCTAGGCAAATGTCCAAAGAACAAGTTGTTCAAAGGCTACATAG
CCCTGGAGCTGCAGCTTCGAGAGTTTGACAGATGCAGGAAGCTTTATGAGAAGTTTCTGGAATTCGGACCTGAAAATTGT
ACCTCGTGGATAAAGTTTGCAGAGTTAGAGACAATCCTTGGTGATGTTGAGAGAGCTCGGGCAATCTATGAGTTAGCCAT
CAGCCAGCCGCGCTTGGATATGCCAGAGCTACTTTGGAAATCCTATATTGATTTTGAGATGGAGCAGGAAGAGCCTGAGA
GAACACGGAACCTTTACCGACGGTTGCTTCAGAGAACACAGCACGTCAAGGTGTGGATCAGTTTGGCTCAGTTTGAGTTC
TCTTCTGGGAAAGAAGGAAGTGTGGCGAAATGCAGACAGATTTATCAAGAAGCCAACAGATCCATGAGACACTGCCAGGA
CAAAGAAGAGAGGCTTATGCTGTTGGAATCCTGGAGAAGTTTTGAAGGGGAGTTTGGAACCGTAGCAGACAAGGAGAGAG
TAGACAGACTCATGCCAGACGAGGTCAAGAAGAGAAGAAAGGTCCAGGCCGACGATGGTTCTGATGCCGGATGGCAAGAA
TACCGGGACTACATCTTTCCAGAGGATGCGGCCCAGCAGCCCAACCTCAAGCTTCTGGCCGTGGCCAAGCTGTGGAAGAA
ACAGCAACAGGAAAGAGAAGCTGCCGGACAGGATCCAGATAAGGACAGGAATGGGAGTGCACTTGCTGGTTAC
ORF - retro_mmus_207 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 88.42 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAASTAAGK-QRIPKVAKVKNKAPAEVQITAEQLLREAKERELELLPPPPQQKITDEEELNDYKLRKRKT |
| MA.ST.A.K.QR.PKVA.VKNKA.AEVQITAEQLLREAKERELELL.PPPQQKITD.EE.NDYKLR.RK. | |
| Retrocopy | MASSTVAKKEQRLPKVAQVKNKAAAEVQITAEQLLREAKERELELLAPPPQQKITDAEEFNDYKLRRRKA |
| Parental | FEDNIRKNRTVISNWIKYAQWEESLKEIQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWD |
| FE.N.R..RTVISNWIKYAQWEESLKE..RARSIYERALDVDYRN.TLWLKYAEMEMKNRQVNHARNIWD | |
| Retrocopy | FEENLRQKRTVISNWIKYAQWEESLKELDRARSIYERALDVDYRNLTLWLKYAEMEMKNRQVNHARNIWD |
| Parental | RAITTLPRVNQFWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQAWHSYINFELRYKEVERARTIYERFV |
| RAITTLPRV.Q.WYKYTYMEE.LGNVAGARQVFERWMEWQPEEQAWHSYIN.ELRYKEVERARTIYERFV | |
| Retrocopy | RAITTLPRVKQLWYKYTYMEELLGNVAGARQVFERWMEWQPEEQAWHSYINLELRYKEVERARTIYERFV |
| Parental | LVHPAVKNWIKYARFEEKHAYFAHARKVYERAVEFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYAL |
| LVHPAVKNWIKYARFEE.HA..AHARKVYERAVEFFG.EHMD.HLYVAFAKFEENQKE.ERVRVIY.YAL | |
| Retrocopy | LVHPAVKNWIKYARFEERHASVAHARKVYERAVEFFGEEHMDQHLYVAFAKFEENQKELERVRVIYRYAL |
| Parental | DRISKQEAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQYEEEVKANPHNYDAWFDYLRLVESDAEADTV |
| DRISKQEAQEL..NYTIFEKKFG.RR..E..IV.KRRF.YEEEVKANPHNYDAWFDYLRLVES.AEADTV | |
| Retrocopy | DRISKQEAQELLTNYTIFEKKFGERRDVEESIVRKRRFHYEEEVKANPHNYDAWFDYLRLVESEAEADTV |
| Parental | REVYERAIANVPPIQEKRHWKRYIYLWVNYALYEELEAKDPERTRQVYQASLELIPHKKFTFAKMWLYYA |
| REVYERAIA.VPP..EKRHWKRYIYLWVNYALYEELEAKDPERTR.VYQASL.LIPH.KFTFAKMWLYYA | |
| Retrocopy | REVYERAIASVPPVLEKRHWKRYIYLWVNYALYEELEAKDPERTRRVYQASLQLIPHTKFTFAKMWLYYA |
| Parental | QFEIRQKNLPFARRALGTSIGKCPKNKLFKGYIELELQLREFDRCRKLYEKFLEFGPENCTSWIKFAELE |
| QFEIRQKNLP.ARRALGTS.GKCPKNKLFKGYI.LELQLREFDRCRKLYEKFLEFGPENCTSWIKFAELE | |
| Retrocopy | QFEIRQKNLPLARRALGTSLGKCPKNKLFKGYIALELQLREFDRCRKLYEKFLEFGPENCTSWIKFAELE |
| Parental | TILGDIERARAIYELAISQPRLDMPEVLWKSYIDFEIEQEETERTRNLYRQLLQRTQHVKVWISFAQFEL |
| TILGD.ERARAIYELAISQPRLDMPE.LWKSYIDFE.EQEE.ERTRNLYR.LLQRTQHVKVWIS.AQFE. | |
| Retrocopy | TILGDVERARAIYELAISQPRLDMPELLWKSYIDFEMEQEEPERTRNLYRRLLQRTQHVKVWISLAQFEF |
| Parental | SSGKEGSVAKCRQIYEEANKTMRNCEEKEERLMLLESWRSFEDEFGTVSDKERVDKLMPEKVKKRRKVQA |
| SSGKEGSVAKCRQIY.EAN..MR.C..KEERLMLLESWRSFE.EFGTV.DKERVD.LMP..VKKRRKVQA | |
| Retrocopy | SSGKEGSVAKCRQIYQEANRSMRHCQDKEERLMLLESWRSFEGEFGTVADKERVDRLMPDEVKKRRKVQA |
| Parental | DDGSDAGWEEYYDYIFPEDAANQPNLKLLAMAKLWKKQQQEREAAEQDPDKDIDESESSSF |
| DDGSDAGW.EY.DYIFPEDAA.QPNLKLLA.AKLWKKQQQEREAA.QDPDKD...S..... | |
| Retrocopy | DDGSDAGWQEYRDYIFPEDAAQQPNLKLLAVAKLWKKQQQEREAAGQDPDKDRNGSALAGY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 15 .30 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 19 .93 RPM |
| SRP007412_heart | 0 .00 RPM | 19 .74 RPM |
| SRP007412_kidney | 0 .00 RPM | 20 .44 RPM |
| SRP007412_liver | 0 .00 RPM | 14 .53 RPM |
| SRP007412_testis | 0 .00 RPM | 42 .95 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_207 was not detected
No EST(s) were mapped for retro_mmus_207 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_123203 | 1038 libraries | 26 libraries | 8 libraries | 0 libraries | 0 libraries |
The graphical summary, for retro_mmus_207 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
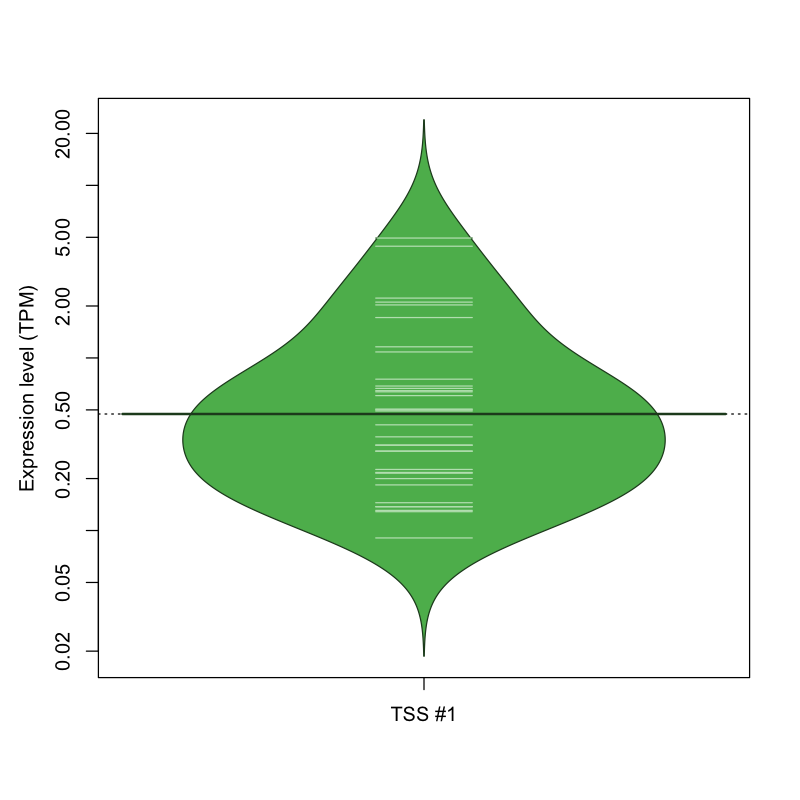
retro_mmus_207 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_207 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 4 parental genes, and 4 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Macropus eugenii | ENSMEUG00000012167 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000016513 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000001767 | 1 retrocopy |
retro_mmus_207 ,
|
| Rattus norvegicus | ENSRNOG00000010587 | 1 retrocopy |