RetrogeneDB ID: | retro_mmus_2710 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 5:148183444..148184389(-) | ||
| Located in intron of: | ENSMUSG00000029651 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000066554 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Fus | ||
| Ensembl ID: | ENSMUSG00000030795 | ||
| Aliases: | Fus, D430004D17Rik, D930039C12Rik, Fus1, Tls | ||
| Description: | fused in sarcoma [Source:MGI Symbol;Acc:MGI:1353633] |

Retrocopy-Parental alignment summary:
>retro_mmus_2710
AATTTTGGGGACATGGCTTCAAACGACTATACCCAACAAGCAACTCAAAGCTACGGGGCCTATCCTACCCAGCCTGGGCA
GGGCTACTCCCAACAGAGCAATCAGCCCTATGGCCAATCAGCTGACACTTCAGGATACGGCCAGATCAGCTATGGTTCCT
CTTCTGGACAGACCCAAAACACAGGCTATGGAACTCAGTCAGCTCCCCAGGGATATGGTTCCACTGGAGGCTATGGCAGC
AGCCAAAGTTCCCAATCTTCTTATGGGCAGCAGTCCTCCTACCCTGGCTATGGCCAACAGCCAGCTCCTAGCAGCACCTC
AGGAAGTTATGGTGGCAGTTCTCAGAGCAGCAGCTATGGGCAACCCCAGAGTTATGGTCAACAGTCTGGCTATGGTGGAC
AGCAACAAAGCTATGGACAACAACAAAGCTCCTATAACCCACCTCAGGGTTATGGACAACAGAACCAGTACAACAGCAGC
AGCGGAGGTGGTGGAGGGGGTGGTGGAGGCAACTATGGCCAAGATCAGTCCTCTATGAGTGGCGGCGGTGGTGGTTATGG
CAATCAGGACTAGAGTGGTGGCGGTGGTGGTGGCTACGGGGGAGGCCAGCAGGATCGTGGGGGCCGTGACAGGGGCAGTG
GAGGTGGTTACAACCGAAGCAGTGGTGGCTATGAACCCAGAGGCTGTGGAGGTAACCGAGGAGGCAGAGGCGTCATGGGC
GGAAGTGACCGTGGTGACTTCAATAAATTTGGTGGTCCTCGGGATCAAGGATCTCGTCATGATTCTGAACAGGATAATTC
AGACAACAATACCATCTTCGTGCAAGGCCTAGGCGAGAATGTTACAATTGAATCTGTGGCTGATTACTTCAAGCAGATTG
GAATTATTAAGACAAACAAGAAAACGGGACAGCCTATGATTAATTTGTACACAGACAGGGAAACT
AATTTTGGGGACATGGCTTCAAACGACTATACCCAACAAGCAACTCAAAGCTACGGGGCCTATCCTACCCAGCCTGGGCA
GGGCTACTCCCAACAGAGCAATCAGCCCTATGGCCAATCAGCTGACACTTCAGGATACGGCCAGATCAGCTATGGTTCCT
CTTCTGGACAGACCCAAAACACAGGCTATGGAACTCAGTCAGCTCCCCAGGGATATGGTTCCACTGGAGGCTATGGCAGC
AGCCAAAGTTCCCAATCTTCTTATGGGCAGCAGTCCTCCTACCCTGGCTATGGCCAACAGCCAGCTCCTAGCAGCACCTC
AGGAAGTTATGGTGGCAGTTCTCAGAGCAGCAGCTATGGGCAACCCCAGAGTTATGGTCAACAGTCTGGCTATGGTGGAC
AGCAACAAAGCTATGGACAACAACAAAGCTCCTATAACCCACCTCAGGGTTATGGACAACAGAACCAGTACAACAGCAGC
AGCGGAGGTGGTGGAGGGGGTGGTGGAGGCAACTATGGCCAAGATCAGTCCTCTATGAGTGGCGGCGGTGGTGGTTATGG
CAATCAGGACTAGAGTGGTGGCGGTGGTGGTGGCTACGGGGGAGGCCAGCAGGATCGTGGGGGCCGTGACAGGGGCAGTG
GAGGTGGTTACAACCGAAGCAGTGGTGGCTATGAACCCAGAGGCTGTGGAGGTAACCGAGGAGGCAGAGGCGTCATGGGC
GGAAGTGACCGTGGTGACTTCAATAAATTTGGTGGTCCTCGGGATCAAGGATCTCGTCATGATTCTGAACAGGATAATTC
AGACAACAATACCATCTTCGTGCAAGGCCTAGGCGAGAATGTTACAATTGAATCTGTGGCTGATTACTTCAAGCAGATTG
GAATTATTAAGACAAACAAGAAAACGGGACAGCCTATGATTAATTTGTACACAGACAGGGAAACT
ORF - retro_mmus_2710 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 88.09 % |
| Parental protein coverage: | 61.58 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | DYTQQATQSYGAYPTQPGQGYSQQSSQPYGQQSYSGYGQSADTSGYGQSSYGSSYGQTQNTGYGTQSAPQ |
| .....A...Y....TQ....Y..Q..Q.Y.QQS...YGQSADTSGYGQ.SYGSS.GQTQNTGYGTQSAPQ | |
| Retrocopy | NFGDMASNDYTQQATQSYGAYPTQPGQGYSQQSNQPYGQSADTSGYGQISYGSSSGQTQNTGYGTQSAPQ |
| Parental | GYGSTGGYGSSQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGGSSQSSSYGQPQSGGYGQQSGYGGQQQSY |
| GYGSTGGYGSSQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGGSSQSSSYGQPQS..YGQQSGYGGQQQSY | |
| Retrocopy | GYGSTGGYGSSQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGGSSQSSSYGQPQS--YGQQSGYGGQQQSY |
| Parental | GQQQSSYNPPQGYGQQNQYNSSSGGGGGGGGGNYGQDQSSMSGGGGGGGYGNQDQSGGGGGGYGGGQQDR |
| GQQQSSYNPPQGYGQQNQYNSSSGGGGGGGGGNYGQDQSSMSGGGGG..YGNQD.SGGGGGGYGGGQQDR | |
| Retrocopy | GQQQSSYNPPQGYGQQNQYNSSSGGGGGGGGGNYGQDQSSMSGGGGG--YGNQD*SGGGGGGYGGGQQDR |
| Parental | GGRGRGGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGPRDQGSRHDSEQDNSDNNTIFVQG |
| GGR.RG.GGGYNRSSGGYEPRG.GG.RGGRG.MGGSDRG.FNKFGGPRDQGSRHDSEQDNSDNNTIFVQG | |
| Retrocopy | GGRDRGSGGGYNRSSGGYEPRGCGGNRGGRGVMGGSDRGDFNKFGGPRDQGSRHDSEQDNSDNNTIFVQG |
| Parental | LGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRET |
| LGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRET | |
| Retrocopy | LGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRET |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 1 .92 RPM | 227 .18 RPM |
| SRP007412_cerebellum | 1 .65 RPM | 283 .28 RPM |
| SRP007412_heart | 1 .81 RPM | 182 .67 RPM |
| SRP007412_kidney | 1 .19 RPM | 174 .92 RPM |
| SRP007412_liver | 0 .49 RPM | 64 .45 RPM |
| SRP007412_testis | 1 .83 RPM | 233 .13 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_2710 was not detected
No EST(s) were mapped for retro_mmus_2710 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
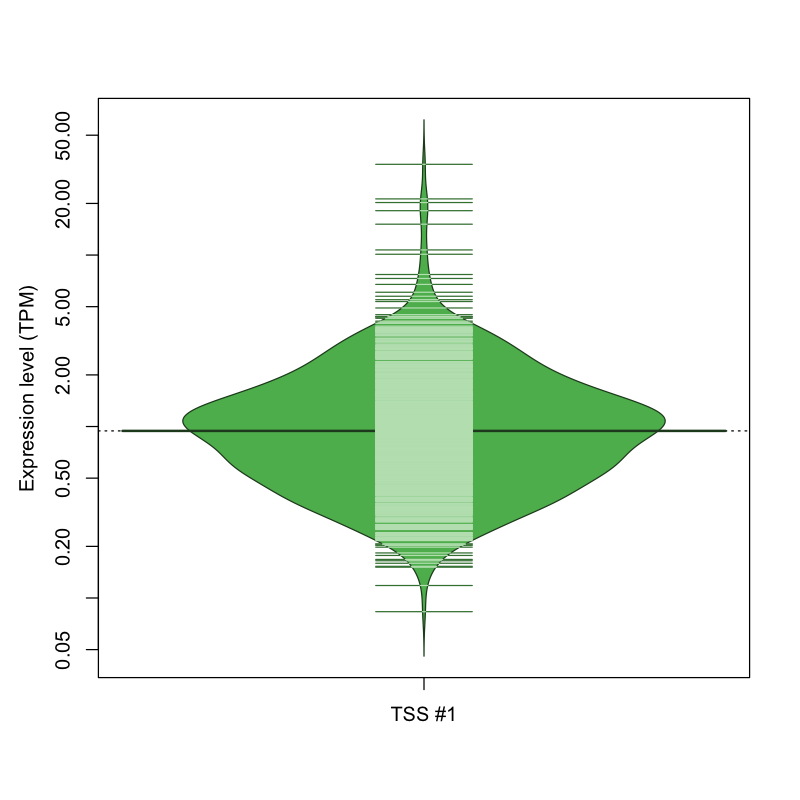
| TSS #1 | TSS_112135 | 235 libraries | 443 libraries | 380 libraries | 7 libraries | 7 libraries |
The graphical summary, for retro_mmus_2710 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2710 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2710 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 6 parental genes, and 12 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Macropus eugenii | ENSMEUG00000000968 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000004806 | 3 retrocopies | |
| Macaca mulatta | ENSMMUG00000019637 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000009079 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000030795 | 2 retrocopies |
retro_mmus_2710 , retro_mmus_2952,
|
| Rattus norvegicus | ENSRNOG00000023360 | 4 retrocopies |