RetrogeneDB ID: | retro_mmus_2855 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 6:92564619..92565840(-) | ||
| Located in intron of: | ENSMUSG00000030020 | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Rbm17 | ||
| Ensembl ID: | ENSMUSG00000037197 | ||
| Aliases: | Rbm17, 2700027J02Rik | ||
| Description: | RNA binding motif protein 17 [Source:MGI Symbol;Acc:MGI:1924188] |

Retrocopy-Parental alignment summary:
>retro_mmus_2855
ATGTCCCTATAGGATGACCTGGGAGTGGAGACCAGTGACTCAAAAACTGAAGGCTGGTCCAAAAGCTTCAAGCTTCTGCA
GTTCCAGCTCCAGGTGAAGAAGGCAGCGCTCACTCAGGCCAAGAGCCAAAGGACCAAGCAAAGTAGGATGCTTGCTCAGG
TCATCAACCTAAAGCGAGGCGGCTCCTCAGATGACCGGCAGATTACAGACACACCACCTCACGTGACAGCTGGGCTGAAG
GACCCTGTGCCCAGTGGGTTTTCTGCAGGGGAAGTTCTGATCCCCTTAGCTGATGAATATGACCCTATGTTCTCTAATGA
CTATGAGAAAGTGGTGAAGCACCAGAGAGAACGGCAGAGGCAGCGGGAGCTGGAAAGACAGAAGGAAATAGAGGAAAGAG
AAAAGAGGCGTAAAGACAGACACGAAGCCAGTAGGCTTTTAAGACAACCAGACCCAGATTCTGATGAGGATAAAGATTAT
GAGCGAGAGCGGAGGAAAAGAAGTATGGGAGGAGCTGCCATCGCCCCACGGAAGTCTCTTGTAGAGAAAGACAAAGAGTT
ACCCTGGGATTTTCCTTATGAAGAGGACTCAAGACCTAGATCACAGTCTTCCAAAGCTGCTATTCCTTCCCCCCCCCCCC
CCATGTATGAGGAGCCAGACAGACCAAGATCTCCAACAGGCCCCAGCAACTCCTTCCTTGCTAACATGGGTGGCACAGTG
GCTCATAAGATTATGCAGAAGTATGGCTTCCGGGAAGGTCAGGGACTGGGGAAACACGAGCAAGGGCTGAGTACTGCATT
GTCTGTGGAGAAGACCAGCAAGCGTGGCGGCAAGATCATTGTGGGGGATGTGAAAGAGAAGGGCAAGGCTCAAGATGCAT
CCAAGAAGTCAGATTCAAATCCATTAACTGAAATTCTTAAGTGTCCTACTAAAGTGGTCTTGCTGAGGAACATGGTTGGT
GCAGGAGAGGTCGATAAAGACTTGGAAGTTGAAACCAAGGAAGAATGTGAAAAATATGGCAAGGTTGGGAAATGTGTGAT
ATTTGAGATTCCTGGTGCCCCTGATGATGAAGCAGTACGGATATTTTTAGAATTTGAGAGAGTAGAATCGGCAATTAAAG
CTGTTGTTGATCTGAATGGGAGGTATTTTGGTGGACAGGTGGTAAAAGCATGTTTCTACAATTTGGATAAATTCAGGGTC
TTGGATCTAGCAGAACAAGTT
ATGTCCCTATAGGATGACCTGGGAGTGGAGACCAGTGACTCAAAAACTGAAGGCTGGTCCAAAAGCTTCAAGCTTCTGCA
GTTCCAGCTCCAGGTGAAGAAGGCAGCGCTCACTCAGGCCAAGAGCCAAAGGACCAAGCAAAGTAGGATGCTTGCTCAGG
TCATCAACCTAAAGCGAGGCGGCTCCTCAGATGACCGGCAGATTACAGACACACCACCTCACGTGACAGCTGGGCTGAAG
GACCCTGTGCCCAGTGGGTTTTCTGCAGGGGAAGTTCTGATCCCCTTAGCTGATGAATATGACCCTATGTTCTCTAATGA
CTATGAGAAAGTGGTGAAGCACCAGAGAGAACGGCAGAGGCAGCGGGAGCTGGAAAGACAGAAGGAAATAGAGGAAAGAG
AAAAGAGGCGTAAAGACAGACACGAAGCCAGTAGGCTTTTAAGACAACCAGACCCAGATTCTGATGAGGATAAAGATTAT
GAGCGAGAGCGGAGGAAAAGAAGTATGGGAGGAGCTGCCATCGCCCCACGGAAGTCTCTTGTAGAGAAAGACAAAGAGTT
ACCCTGGGATTTTCCTTATGAAGAGGACTCAAGACCTAGATCACAGTCTTCCAAAGCTGCTATTCCTTCCCCCCCCCCCC
CCATGTATGAGGAGCCAGACAGACCAAGATCTCCAACAGGCCCCAGCAACTCCTTCCTTGCTAACATGGGTGGCACAGTG
GCTCATAAGATTATGCAGAAGTATGGCTTCCGGGAAGGTCAGGGACTGGGGAAACACGAGCAAGGGCTGAGTACTGCATT
GTCTGTGGAGAAGACCAGCAAGCGTGGCGGCAAGATCATTGTGGGGGATGTGAAAGAGAAGGGCAAGGCTCAAGATGCAT
CCAAGAAGTCAGATTCAAATCCATTAACTGAAATTCTTAAGTGTCCTACTAAAGTGGTCTTGCTGAGGAACATGGTTGGT
GCAGGAGAGGTCGATAAAGACTTGGAAGTTGAAACCAAGGAAGAATGTGAAAAATATGGCAAGGTTGGGAAATGTGTGAT
ATTTGAGATTCCTGGTGCCCCTGATGATGAAGCAGTACGGATATTTTTAGAATTTGAGAGAGTAGAATCGGCAATTAAAG
CTGTTGTTGATCTGAATGGGAGGTATTTTGGTGGACAGGTGGTAAAAGCATGTTTCTACAATTTGGATAAATTCAGGGTC
TTGGATCTAGCAGAACAAGTT
ORF - retro_mmus_2855 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 92.4 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSLYDDLGVETSDSKTEGWSKNFKLLQSQLQVKKAALTQAKSQRTKQSTVLAPVIDLKRGGSSDDRQIAD |
| MSL.DDLGVETSDSKTEGWSK.FKLLQ.QLQVKKAALTQAKSQRTKQS..LA.VI.LKRGGSSDDRQI.D | |
| Retrocopy | MSL*DDLGVETSDSKTEGWSKSFKLLQFQLQVKKAALTQAKSQRTKQSRMLAQVINLKRGGSSDDRQITD |
| Parental | TPPHVAAGLKDPVPSGFSAGEVLIPLADEYDPMFPNDYEKVVKRQREERQRQRELERQKEIEEREKRRKD |
| TPPHV.AGLKDPVPSGFSAGEVLIPLADEYDPMF.NDYEKVVK.QRE.RQRQRELERQKEIEEREKRRKD | |
| Retrocopy | TPPHVTAGLKDPVPSGFSAGEVLIPLADEYDPMFSNDYEKVVKHQRE-RQRQRELERQKEIEEREKRRKD |
| Parental | RHEASGFSRRPDPDSDEDEDYERERRKRSMGGAAIAPPTSLVEKDKELPRDFPYEEDSRPRSQSSKAAIP |
| RHEAS...R.PDPDSDED.DYERERRKRSMGGAAIAP..SLVEKDKELP.DFPYEEDSRPRSQSSKAAIP | |
| Retrocopy | RHEASRLLRQPDPDSDEDKDYERERRKRSMGGAAIAPRKSLVEKDKELPWDFPYEEDSRPRSQSSKAAIP |
| Parental | PP---VYEEPDRPRSPTGPSNSFLANMGGTVAHKIMQKYGFREGQGLGKHEQGLSTALSVEKTSKRGGKI |
| ......YEEPDRPRSPTGPSNSFLANMGGTVAHKIMQKYGFREGQGLGKHEQGLSTALSVEKTSKRGGKI | |
| Retrocopy | XXXXXMYEEPDRPRSPTGPSNSFLANMGGTVAHKIMQKYGFREGQGLGKHEQGLSTALSVEKTSKRGGKI |
| Parental | IVGDATEKGEAQDASKKSDSNPLTEILKCPTKVVLLRNMVGAGEVDEDLEVETKEECEKYGKVGKCVIFE |
| IVGD..EKG.AQDASKKSDSNPLTEILKCPTKVVLLRNMVGAGEVD.DLEVETKEECEKYGKVGKCVIFE | |
| Retrocopy | IVGDVKEKGKAQDASKKSDSNPLTEILKCPTKVVLLRNMVGAGEVDKDLEVETKEECEKYGKVGKCVIFE |
| Parental | IPGAPDDEAVRIFLEFERVESAIKAVVDLNGRYFGGRVVKACFYNLDKFRVLDLAEQV |
| IPGAPDDEAVRIFLEFERVESAIKAVVDLNGRYFGG.VVKACFYNLDKFRVLDLAEQV | |
| Retrocopy | IPGAPDDEAVRIFLEFERVESAIKAVVDLNGRYFGGQVVKACFYNLDKFRVLDLAEQV |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 30 .96 RPM |
| SRP007412_cerebellum | 0 .26 RPM | 61 .27 RPM |
| SRP007412_heart | 0 .00 RPM | 31 .01 RPM |
| SRP007412_kidney | 0 .29 RPM | 48 .51 RPM |
| SRP007412_liver | 0 .16 RPM | 48 .58 RPM |
| SRP007412_testis | 2 .93 RPM | 46 .20 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_2855 was not detected
No EST(s) were mapped for retro_mmus_2855 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
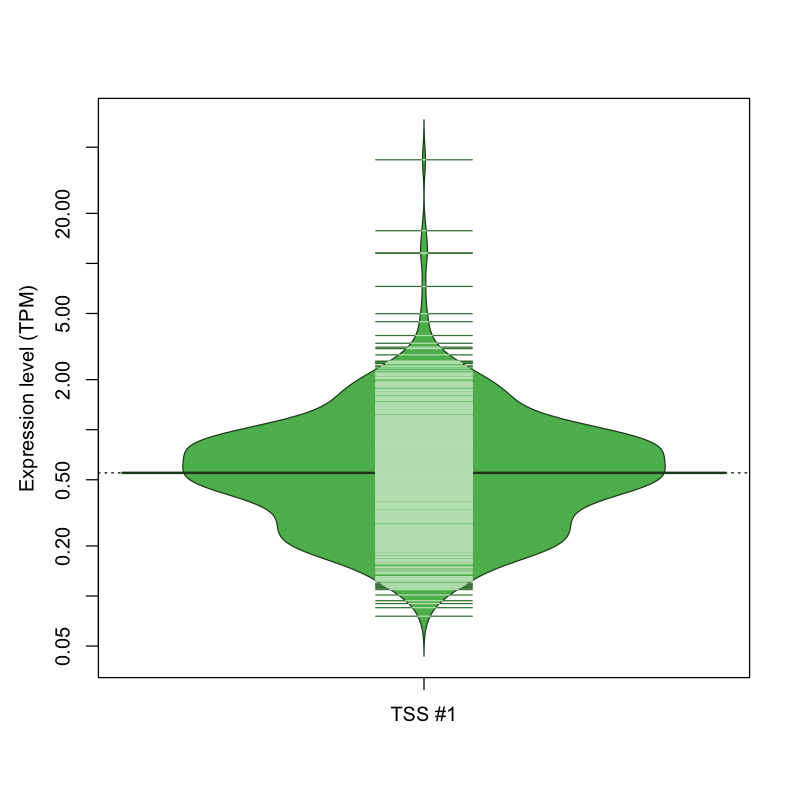
| TSS #1 | TSS_125940 | 385 libraries | 551 libraries | 131 libraries | 1 library | 4 libraries |
The graphical summary, for retro_mmus_2855 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2855 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2855 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 14 parental genes, and 16 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000021951 | 2 retrocopies | |
| Canis familiaris | ENSCAFG00000005206 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000020573 | 1 retrocopy | |
| Equus caballus | ENSECAG00000008939 | 1 retrocopy | |
| Erinaceus europaeus | ENSEEUG00000013077 | 1 retrocopy | |
| Homo sapiens | ENSG00000134453 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000011347 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000015152 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000037197 | 1 retrocopy |
retro_mmus_2855 ,
|
| Otolemur garnettii | ENSOGAG00000015687 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000002067 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000002265 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000013591 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000001344 | 1 retrocopy |