RetrogeneDB ID: | retro_mmus_3475 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:75915490..75916523(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000083544 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Oat | ||
| Ensembl ID: | ENSMUSG00000030934 | ||
| Aliases: | Oat, AI194874 | ||
| Description: | ornithine aminotransferase [Source:MGI Symbol;Acc:MGI:97394] |

Retrocopy-Parental alignment summary:
>retro_mmus_3475
AAGATCATAGATGCTGTGAAGAGTCAGGTGGACAAGTCAACATTAATATCTCAGGCTTTCTATAACTATGTCCTTGGTGA
ATACAAGCAGTACATCACCAAGCTTTTCAACTACAACAAAGTTCTTCCTATGAATACAGGAGTGGAGGCTGGAGAGACTG
AATGTAAGCTTTCTTGTCCTTGGGCTACACAGTGAAAGGCATCCAGAAATACAAAGCAAAGATTTTTTTTTTCTGTTGGA
AACGTTTGGGGTTGAAAGTTGTGAGCAGTCTCCAGTTCTGCAGATCCGTCCAGTTACAATGGCTTTGGATTCTTCCTGCC
AGGCTTTGAAACCATCCCGTTTAAGGATCTGCCTACACTGGAGCATGCTCTTCAGGATCCAAATGTTGCTGCTTTCATGG
TGGGGCCCATCCAGGGTGAAGCAGGTGTTATTGTTCTGGATCCAAGACACCTGACCAGAGTTTGAGAACTCTGTACCAGG
CACCAGGTTCTGTTTATTGCTGATGAAATACAGACAGGATTGGCCAAAACTGGTAGATGGCTAGCTGTGGATCATAAAAA
TTTCAGACCTGACATAGTTCTTCTTGGGAAGGCCCTTTCTGATGGCTTATACCCTTTGTCTGCAGTCCTGTGTGATGATG
ACATAACGCTGACCAATAAGACAGGCGAGCATGGCTCTACATACGGCAGCAACCCACTAGGCAGCCAAATTGCCATTGTG
GTTCTTGAGGTTTTAGAAGAAGATAATCTTGCTGAAAATGCAGACAAGATAGGTGCTATCTTGAGAAAGGAGCTCACGAA
GCTGCCCTCTGACCTTGTGACTGCTGTGAGAGAGAAAGGGTTGCTAAATGCCATTGTCATCAGAGAAAACAAAGATTGTG
ATACTTGGATGGTTGTGCCTGCAACTCTGAGATAATGGGCTTCTGGCCAAGCCAACTCACTGCGATATCATCAGGTTTGC
CTCTCCACTTGTGATCAAGGAAGATGAGATCCAGGAGTCAGTGGAGATCATCAACAAGGCCGTCTTGTTCTTC
ORF - retro_mmus_3475 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 78.96 % |
| Parental protein coverage: | 78.36 % |
| Number of stop codons detected: | 4 |
| Number of frameshifts detected: | 3 |
Retrocopy - Parental Gene Alignment:
| Parental | KIIDAMKSQVDKLTLTSRAFYNNVLGEYEEYITKLFNYNKVLPMNTGVEAGETACKLARRW-GYTVKGIQ |
| KIIDA.KSQVDK.TL.S.AFYN.VLGEY..YITKLFNYNKVLPMNTGVEAGET.CKL...W.GYTVKGIQ | |
| Retrocopy | KIIDAVKSQVDKSTLISQAFYNYVLGEYKQYITKLFNYNKVLPMNTGVEAGETECKLSCPW<GYTVKGIQ |
| Parental | KYKA-KIVFADGNFWGRTLSAISSSTDPTSYDGFGPFMPGFETIPYNDLPALERALQDPNVAAFMVEPIQ |
| KYKA.......GN.WG..L.A.SSS.DP.SY.GFG.F.PGFETIP..DLP.LE.ALQDPNVAAFMV.PIQ | |
| Retrocopy | KYKA>RXXXXVGNVWG*KL*AVSSSADPSSYNGFGFFLPGFETIPFKDLPTLEHALQDPNVAAFMVGPIQ |
| Parental | GEAGVIVPDPGYLTGVRELCTRHQVLFIADEIQTGLARTGRWLAVDHENVRPDMVLLGKALSGGLYPVSA |
| GEAGVIV.DP..LT.V.ELCTRHQVLFIADEIQTGLA.TGRWLAVDH.N.RPD.VLLGKALS.GLYP.SA | |
| Retrocopy | GEAGVIVLDPRHLTRV*ELCTRHQVLFIADEIQTGLAKTGRWLAVDHKNFRPDIVLLGKALSDGLYPLSA |
| Parental | VLCDDEIMLTIKPGEHGSTYGGNPLGCRIAIAALEVLEEENLAENADKMGAILRKELMKLPSDVVTSVRG |
| VLCDD.I.LT.K.GEHGSTYG.NPLG..IAI..LEVLEE.NLAENADK.GAILRKEL.KLPSD.VT.VR. | |
| Retrocopy | VLCDDDITLTNKTGEHGSTYGSNPLGSQIAIVVLEVLEEDNLAENADKIGAILRKELTKLPSDLVTAVRE |
| Parental | KGLLNAIVIRETKDCDAWKV-CLRLRDNGLLAKPTHGDIIRLAPPLVIKEDEIRESVEIINKTILSF |
| KGLLNAIVIRE.KDCD.W.V.CL.L.DNGLLAKPTH.DIIR.A.PLVIKEDEI.ESVEIINK..L.F | |
| Retrocopy | KGLLNAIVIRENKDCDTWMV>CLQL*DNGLLAKPTHCDIIRFASPLVIKEDEIQESVEIINKAVLFF |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 67 .03 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 62 .35 RPM |
| SRP007412_heart | 0 .00 RPM | 146 .03 RPM |
| SRP007412_kidney | 0 .02 RPM | 213 .19 RPM |
| SRP007412_liver | 0 .05 RPM | 678 .15 RPM |
| SRP007412_testis | 0 .00 RPM | 25 .25 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_3475 was not detected
No EST(s) were mapped for retro_mmus_3475 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_158062 | 372 libraries | 247 libraries | 365 libraries | 61 libraries | 27 libraries |
The graphical summary, for retro_mmus_3475 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
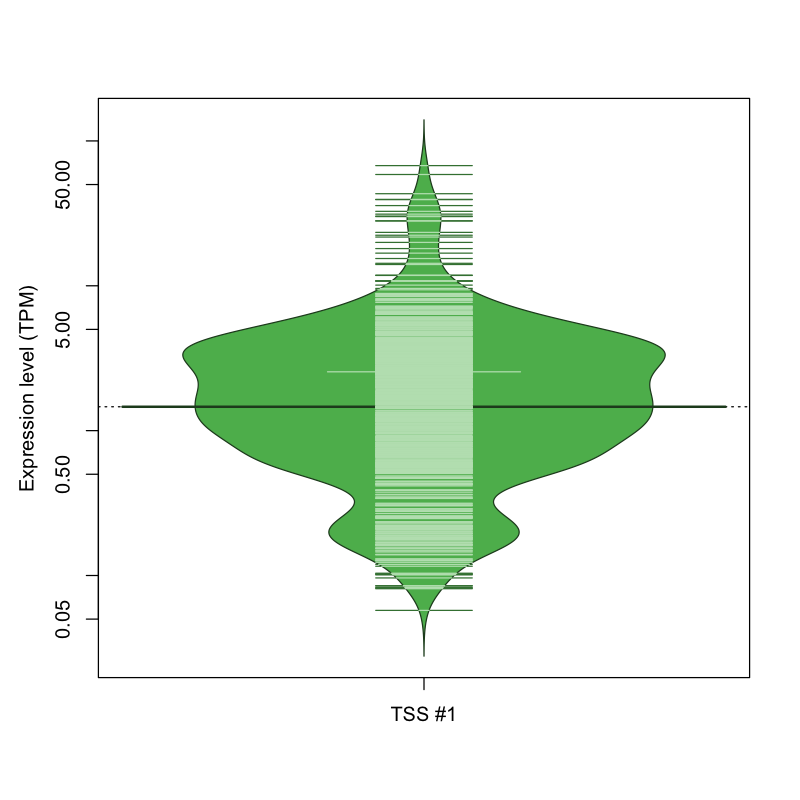
retro_mmus_3475 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_3475 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 10 parental genes, and 12 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000012633 | 1 retrocopy | |
| Homo sapiens | ENSG00000065154 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000000848 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000005655 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000030934 | 1 retrocopy |
retro_mmus_3475 ,
|
| Nomascus leucogenys | ENSNLEG00000000482 | 2 retrocopies | |
| Pongo abelii | ENSPPYG00000002737 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000003028 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000010734 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000004225 | 1 retrocopy |