RetrogeneDB ID: | retro_hsap_41 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 4:96761301..96762468(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000163114 | |
| Aliases: | PDHA2, PDHAL | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | PDHA1 | ||
| Ensembl ID: | ENSG00000131828 | ||
| Aliases: | PDHA1, PDHA, PDHCE1A, PHE1A | ||
| Description: | pyruvate dehydrogenase (lipoamide) alpha 1 [Source:HGNC Symbol;Acc:8806] |

Retrocopy-Parental alignment summary:
>retro_hsap_41
ATGCTGGCCGCCTTCATCTCCCGCGTGTTGAGGCGAGTTGCCCAGAAATCAGCTCGCAGAGTGCTGGTGGCATCCCGTAA
CTCCTCAAATGACGCTACATTTGAAATTAAGAAATGTGATCTTTATCTGTTGGAAGAGGGTCCCCCTGTCACTACAGTGC
TCACTAGGGCGGAGGGGCTTAAATACTACAGGATGATGCTGACTGTTCGCCGCATGGAATTGAAGGCAGATCAGCTGTAC
AAACAGAAATTCATTCGCGGTTTCTGTCACCTGTGCGATGGTCAGGAAGCTTGTTGCGTGGGCCTTGAGGCCGGCATAAA
CCCCTCGGATCACGTCATTACATCCTATAGGGCTCATGGTGTGTGCTATACTCGGGGACTTTCTGTCCGATCCATTCTCG
CAGAGCTGACGGGAAGAAGAGGAGGTTGTGCTAAAGGAAAAGGAGGATCGATGCATATGTATACCAAGAACTTCTATGGG
GGCAATGGCATCGTCGGTGCACAGGGCCCCCTGGGCGCTGGCATTGCTCTGGCCTGTAAATATAAAGGAAACGATGAGAT
CTGTTTGACTTTATATGGGGATGGCGCTGCGAATCAGGGGCAGATAGCCGAAGCTTTCAATATGGCAGCTTTATGGAAAT
TACCTTGTGTTTTCATCTGTGAGAATAACCTATATGGAATGGGAACATCTACTGAGAGAGCAGCAGCCAGCCCTGATTAC
TACAAGAGGGGCAATTTTATCCCTGGGCTAAAGGTCGATGGAATGGATGTTCTGTGTGTTCGTGAGGCAACAAAATTTGC
AGCTAACTACTGTAGATCTGGAAAGGGGCCCATACTGATGGAGCTGCAAACCTACCGTTATCATGGACACAGTATGAGTG
ATCCTGGAGTCAGTTATCGTACACGAGAAGAAATTCAGGAAGTAAGAAGTAAGAGGGATCCTATAATAATTCTCCAAGAT
AGAATGGTAAACAGCAAGCTCGCCACTGTGGAAGAATTAAAGGAAATTGGGGCTGAGGTGAGGAAAGAAATTGATGATGC
TGCCCAGTTTGCTACCACTGATCCTGAGCCACATTTGGAAGAATTAGGCCATCACATCTACAGCAGTGATTCATCTTTTG
AAGTTCGTGGTGCAAATCCATGGATCAAGTTTAAGTCCGTCAGTTAA
ATGCTGGCCGCCTTCATCTCCCGCGTGTTGAGGCGAGTTGCCCAGAAATCAGCTCGCAGAGTGCTGGTGGCATCCCGTAA
CTCCTCAAATGACGCTACATTTGAAATTAAGAAATGTGATCTTTATCTGTTGGAAGAGGGTCCCCCTGTCACTACAGTGC
TCACTAGGGCGGAGGGGCTTAAATACTACAGGATGATGCTGACTGTTCGCCGCATGGAATTGAAGGCAGATCAGCTGTAC
AAACAGAAATTCATTCGCGGTTTCTGTCACCTGTGCGATGGTCAGGAAGCTTGTTGCGTGGGCCTTGAGGCCGGCATAAA
CCCCTCGGATCACGTCATTACATCCTATAGGGCTCATGGTGTGTGCTATACTCGGGGACTTTCTGTCCGATCCATTCTCG
CAGAGCTGACGGGAAGAAGAGGAGGTTGTGCTAAAGGAAAAGGAGGATCGATGCATATGTATACCAAGAACTTCTATGGG
GGCAATGGCATCGTCGGTGCACAGGGCCCCCTGGGCGCTGGCATTGCTCTGGCCTGTAAATATAAAGGAAACGATGAGAT
CTGTTTGACTTTATATGGGGATGGCGCTGCGAATCAGGGGCAGATAGCCGAAGCTTTCAATATGGCAGCTTTATGGAAAT
TACCTTGTGTTTTCATCTGTGAGAATAACCTATATGGAATGGGAACATCTACTGAGAGAGCAGCAGCCAGCCCTGATTAC
TACAAGAGGGGCAATTTTATCCCTGGGCTAAAGGTCGATGGAATGGATGTTCTGTGTGTTCGTGAGGCAACAAAATTTGC
AGCTAACTACTGTAGATCTGGAAAGGGGCCCATACTGATGGAGCTGCAAACCTACCGTTATCATGGACACAGTATGAGTG
ATCCTGGAGTCAGTTATCGTACACGAGAAGAAATTCAGGAAGTAAGAAGTAAGAGGGATCCTATAATAATTCTCCAAGAT
AGAATGGTAAACAGCAAGCTCGCCACTGTGGAAGAATTAAAGGAAATTGGGGCTGAGGTGAGGAAAGAAATTGATGATGC
TGCCCAGTTTGCTACCACTGATCCTGAGCCACATTTGGAAGAATTAGGCCATCACATCTACAGCAGTGATTCATCTTTTG
AAGTTCGTGGTGCAAATCCATGGATCAAGTTTAAGTCCGTCAGTTAA
ORF - retro_hsap_41 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 85.82 % |
| Parental protein coverage: | 99.23 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MLAA-VSRVLSGASQKPASRVLVASRNFANDATFEIKKCDLHRLEEGPPVTTVLTREDGLKYYRMMQTVR |
| MLAA..SRVL....QK.A.RVLVASRN..NDATFEIKKCDL..LEEGPPVTTVLTR..GLKYYRMM.TVR | |
| Retrocopy | MLAAFISRVLRRVAQKSARRVLVASRNSSNDATFEIKKCDLYLLEEGPPVTTVLTRAEGLKYYRMMLTVR |
| Parental | RMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGINPTDHLITAYRAHGFTFTRGLSVREILAELTGRK |
| RMELKADQLYKQK.IRGFCHLCDGQEACCVGLEAGINP.DH.IT.YRAHG...TRGLSVR.ILAELTGR. | |
| Retrocopy | RMELKADQLYKQKFIRGFCHLCDGQEACCVGLEAGINPSDHVITSYRAHGVCYTRGLSVRSILAELTGRR |
| Parental | GGCAKGKGGSMHMYAKNFYGGNGIVGAQVPLGAGIALACKYNGKDEVCLTLYGDGAANQGQIFEAYNMAA |
| GGCAKGKGGSMHMY.KNFYGGNGIVGAQ.PLGAGIALACKY.G.DE.CLTLYGDGAANQGQI.EA.NMAA | |
| Retrocopy | GGCAKGKGGSMHMYTKNFYGGNGIVGAQGPLGAGIALACKYKGNDEICLTLYGDGAANQGQIAEAFNMAA |
| Parental | LWKLPCIFICENNRYGMGTSVERAAASTDYYKRGDFIPGLRVDGMDILCVREATRFAAAYCRSGKGPILM |
| LWKLPC.FICENN.YGMGTS.ERAAAS.DYYKRG.FIPGL.VDGMD.LCVREAT.FAA.YCRSGKGPILM | |
| Retrocopy | LWKLPCVFICENNLYGMGTSTERAAASPDYYKRGNFIPGLKVDGMDVLCVREATKFAANYCRSGKGPILM |
| Parental | ELQTYRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNSNLASVEELKEIDVEVRKEIEDAAQF |
| ELQTYRYHGHSMSDPGVSYRTREEIQEVRSK.DPI..L.DRMVNS.LA.VEELKEI..EVRKEI.DAAQF | |
| Retrocopy | ELQTYRYHGHSMSDPGVSYRTREEIQEVRSKRDPIIILQDRMVNSKLATVEELKEIGAEVRKEIDDAAQF |
| Parental | ATADPEPPLEELGYHIYSSDPPFEVRGANQWIKFKSVS |
| AT.DPEP.LEELG.HIYSSD..FEVRGAN.WIKFKSVS | |
| Retrocopy | ATTDPEPHLEELGHHIYSSDSSFEVRGANPWIKFKSVS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 51 .13 RPM |
| bodymap2_adrenal | 0 .04 RPM | 51 .81 RPM |
| bodymap2_brain | 0 .00 RPM | 61 .46 RPM |
| bodymap2_breast | 0 .00 RPM | 102 .47 RPM |
| bodymap2_colon | 0 .00 RPM | 51 .43 RPM |
| bodymap2_heart | 0 .00 RPM | 238 .15 RPM |
| bodymap2_kidney | 0 .00 RPM | 149 .42 RPM |
| bodymap2_liver | 0 .00 RPM | 52 .60 RPM |
| bodymap2_lung | 0 .00 RPM | 34 .65 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 38 .81 RPM |
| bodymap2_ovary | 0 .00 RPM | 50 .99 RPM |
| bodymap2_prostate | 0 .00 RPM | 61 .04 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 97 .80 RPM |
| bodymap2_testis | 68 .57 RPM | 37 .59 RPM |
| bodymap2_thyroid | 0 .00 RPM | 87 .90 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 31 .93 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_41 was not detected
10 EST(s) were mapped to retro_hsap_41 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AI393336 | 96761616 | 96761833 | 100 | 217 | 0 | 217 |
| BU854475 | 96761264 | 96761763 | 95.2 | 488 | 11 | 470 |
| DB065822 | 96761278 | 96761860 | 100 | 582 | 0 | 582 |
| DB459674 | 96761243 | 96761875 | 100 | 631 | 0 | 631 |
| DB461656 | 96761274 | 96761745 | 100 | 471 | 0 | 471 |
| HY026399 | 96761255 | 96761691 | 99.8 | 435 | 1 | 434 |
| HY029492 | 96761264 | 96761736 | 100 | 472 | 0 | 472 |
| HY048052 | 96761274 | 96761768 | 99.8 | 493 | 1 | 492 |
| HY051642 | 96761248 | 96761678 | 99.6 | 428 | 2 | 426 |
| HY053842 | 96761237 | 96761715 | 99.8 | 477 | 1 | 476 |
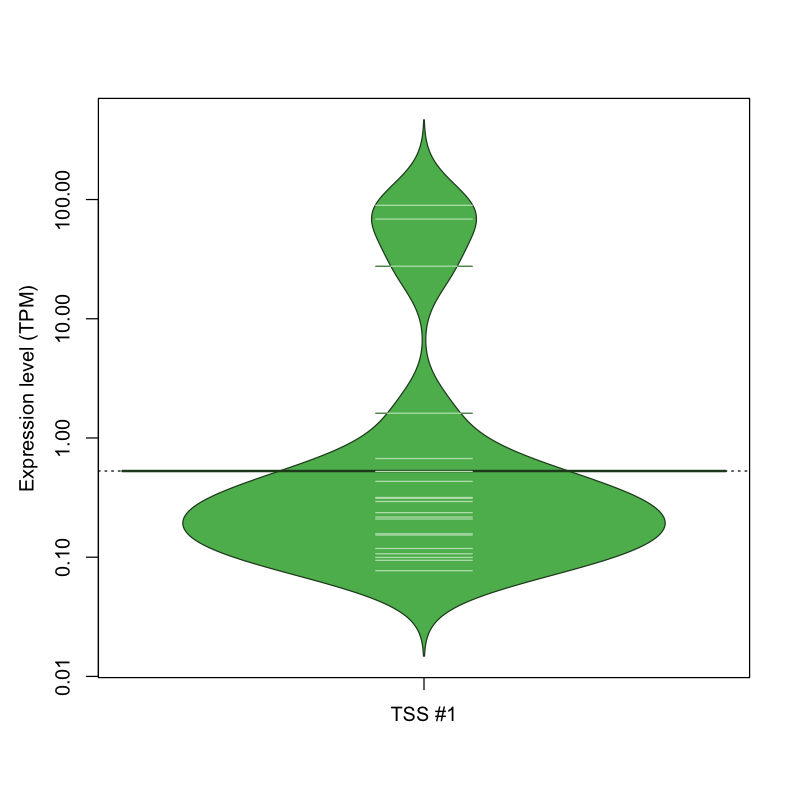
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_147105 | 1809 libraries | 16 libraries | 1 library | 0 libraries | 3 libraries |
The graphical summary, for retro_hsap_41 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_41 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_41 has 9 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 22 parental genes, and 27 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .00 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .00 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .03 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .03 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .00 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .03 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .02 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .02 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .00 RPM |