
Help contents:
i. Browse/Search record
ii. Unclassified
iii. RNA-Seq
Browse/Search record
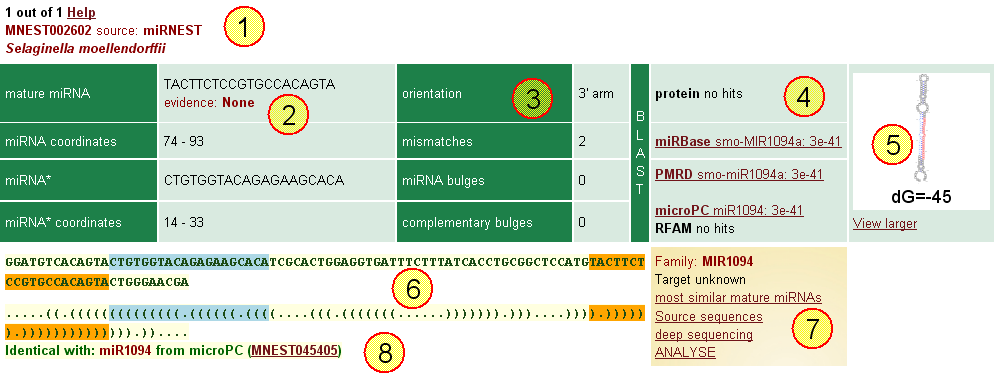 Fig. 1 An example record for plant miRNA.
Fig. 1 An example record for plant miRNA.
![]() MNEST002602: miRNEST database accession number. Having the accession number you can find the record here.
MNEST002602: miRNEST database accession number. Having the accession number you can find the record here.
![]() Some basic information on predicted miRNA: sequence of mature miRNA, miRNA* and their start-end coordinates on pre-miRNA. Also, evidence information is provided here.
Some basic information on predicted miRNA: sequence of mature miRNA, miRNA* and their start-end coordinates on pre-miRNA. Also, evidence information is provided here.
![]() If orientation is up/down, it means that mature miRNA is located in the upper/lower arm of hairpin structure.
If orientation is up/down, it means that mature miRNA is located in the upper/lower arm of hairpin structure.
Mismatches, unpaired bases in RNA secondary structure, were counted for the hairpin region that is occupied by mature miRNA. Wobble (G:U) pairs are not considered as mismatches here.
Similarly, miRNA bulges means the number of bulges present within the coordinates of mature miRNA and on the same hairpin arm, while complementary bulges were calculated for the opposite arm.
![]() In this section, BLAST results are shown. Here, pre-miRNA as Query was used and the databases were:
In this section, BLAST results are shown. Here, pre-miRNA as Query was used and the databases were:
- UniProt
- miRBase Release 16
- PMRD (Plant microRNA Database)
- microPC
- RFAM
BLAST results can be viewed in alignment format by clicking the links.
![]() pre-miRNA secondary structure, as predicted by RNAfold can be viewed and edited in VARNA (Java Applet). The folding free energy is provided at the bottom. Also, there is a possibility to see the structure in large view. In both small and large view, mature miRNA is marked with orange and miRNA* with lightblue.
pre-miRNA secondary structure, as predicted by RNAfold can be viewed and edited in VARNA (Java Applet). The folding free energy is provided at the bottom. Also, there is a possibility to see the structure in large view. In both small and large view, mature miRNA is marked with orange and miRNA* with lightblue.
![]() In this section, pre-miRNA sequence and dot-plot representation of secondary structure (by RNAfold) are shown. Mature miRNA is highlighted in orange, and miRNA* - in lightblue.
In this section, pre-miRNA sequence and dot-plot representation of secondary structure (by RNAfold) are shown. Mature miRNA is highlighted in orange, and miRNA* - in lightblue.
![]() The components of this section are as follows:
The components of this section are as follows:
- miRNA candidate assignment to miRNA family, according the nomenclature and annotations from miRBase Release 16 and PMRD. In the example above the family name is MIRfam1094 (PMRD)
- targets information: our predictions as well as data from miRDB, PMRD, miRTarBase, miRecords abd ASRP.
- most similar mature miRNAs - from miRBase and PMRD - with up to 4 mismatches when compared to the mature miRNA of interest.
- For each record out of miRNEST predictions, source contig sequence is provided. It is also possible to reach all the EST sequences, that were assembled into the contig.
- deep sequencing link leads the information on small RNA seep sequencing reads that were mapped to the pre-miRNA.
- additional data gives access to data from external resources: miRBase, dPORE-miRNA, PhenomiR, Patrocles, dbDEMC, CoGemiR, PMRD, and ncRNA imprint.
- upon clicking ANALYSE, there is a possibility to run BLASTN against microRNAs from miRBase, PMRD, microPC, miRNEST candidates and miRNA candidates by Huang et al. and Hao et al. The user has a chance to select cutoff values for wordsize, E-value as well as select output type. It is also possible to run ClustalW analysis, but first BLASTN parametres for sequence clustering need to be selected.
![]() This indicates miRNEST records that have identical pre-mirNA sequence, yet there might be a difference in sequence length.
This indicates miRNEST records that have identical pre-mirNA sequence, yet there might be a difference in sequence length.
Unclassified
The records belonging to this section generally have a very similar format to the one in Browse section.
It is divided into two parts:
a) plant and animal candidates showing E-value of 1e-20 or lower for BLASTX against UniProt proteins.
b) animal candidates with pre-miRNA length exceeding 215 bases. None of known animal miRNAs from miRBase (Release 17) exceeds this length limit, however some of the candidates show quite a high similarity
to known miRNAs.
RNA-seq
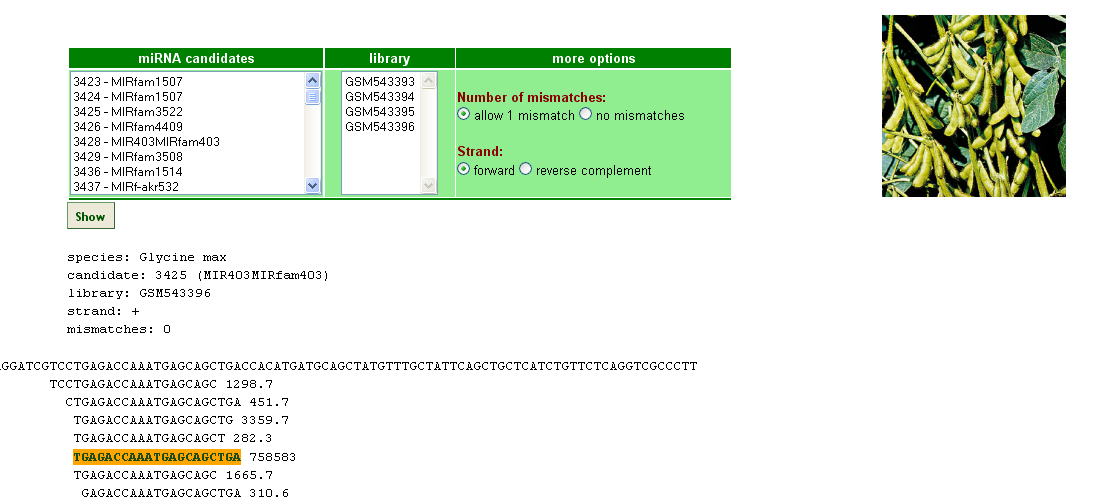
Fig. 2 A sample RNA-Seq record.
This section serves to visualize the results of mapping of reads from small RNA deep sequencing libraries to predicted pre-miRNAs.
It is possible to choose whether mismatches will be allowed or not. Mismatches are marked with lightblue.
Mature miRNA is marked with orange and miRNA* with lightblue.
Mismatched nucleotides are in small capitals and marked with lightgreen.
Go to:
i. Browse
ii. Unclassified
iii. RNA-Seq