RetrogeneDB ID: | retro_hsap_1451 | ||
Retrocopy location | Organism: | Human (Homo sapiens) | |
| Coordinates: | 14:89756688..89757700(-) | ||
| Located in intron of: | ENSG00000053254 | ||
Retrocopy information | Ensembl ID: | ENSG00000259151 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | CAP2 | ||
| Ensembl ID: | ENSG00000112186 | ||
| Aliases: | None | ||
| Description: | CAP, adenylate cyclase-associated protein, 2 (yeast) [Source:HGNC Symbol;Acc:20039] |

Retrocopy-Parental alignment summary:
>retro_hsap_1451
AGGGCTTTCCTTCTGATGGCATCTCATGACCAACAACCCCGAGAGAATGCGTGGCTGCATTTCTGAAACCCAAATCAGAA
AAGATTCAGAAGTCCAAACCTTCAAAGAGAGAAACCGAGGGAGTAACATGTTTAGTCATCTCTCGGATGTCAGCAAAAGC
CTCCCTGCCCTTGGCTGCATAACTGTGCCCCCCAACTTGGGTACTTACGTCAACGAGATGTGTGATGCTGCCACCTTTTA
CACTAACAGGGTCTTAAAGGGCTACAAACATGATTTACACCGTGTGGATTGGGTGAGGTCATATGTGAACATTTGGAGTG
AGTTTCAAGCATGAGTCAAGGAGCACCACACCACGGGCCTCACGTGGAGCAGAACAGGTCCGCAGCACCCACATCAGCGC
TTTCTGTTCTCTCCTGAGGGCCTGACCTTTCTCCACATCCTCCTCACCTCCTCCAGGGCCCCTCCACTTTTTGAGAATGA
AGACACAAAAGAAAAGTCCTCTCCTTCATGCTCAGCTTTATTTGCCCGGTTTAACCAGGGAGAAGCCTAACCCTGCAGAA
CCAGGGCTCTGGCACATCACTGATGACCAGAAGGCCTATATAAGAATCCCAGCCTGCAGGCTCACGGAGGGCAAAGTACG
TCTCCCCACAGAAGCTACATTCCAAGTCCCACATCTCCCAAACCTCATCCTTCCCAGCAACATGCACCCGTGTTCAAGTT
GGAAGGAAGTAAATGGAGAGCAGAGTACCAAGAAGACAGGAATGACCTTGTGATTTCAGAGACTGAGCTGAAACAAGTGG
CTTACATTTTCAATTGTGAAAAATCAGCTCTCCCGATGAAAGGGAAAGTAAACTCCATTCCAGTGGAAAACTGTAAGAAG
TTTGGTCTGGTCTTTGACAGTTTGGTGGGCATTGGGGAAATGACCAACTCCAAGTACATTCAAATCCAGGTAATGGGGAG
AGTACCAGCAATTTCCACTAATAAGACAGAAGGTTGCCACACATACCTAACT
ORF - retro_hsap_1451 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 71.18 % |
| Parental protein coverage: | 71.07 % |
| Number of stop codons detected: | 5 |
| Number of frameshifts detected: | 5 |
Retrocopy - Parental Gene Alignment:
| Parental | RAFLLMASQYQQPHEND-VAALLKPISEKIQ-EIQTFRERNRGSNMFNHLSAVSESIPALGWIAVSPKPG |
| RAFLLMAS..QQP.EN..VAA.LKP.SEKIQ.E.QTF.ERNRGSNMF.HLS.VS.S.PALG.I.V.P..G | |
| Retrocopy | RAFLLMASHDQQPRENA<VAAFLKPKSEKIQ<EVQTFKERNRGSNMFSHLSDVSKSLPALGCITVPPNLG |
| Parental | PYVKEMNDAATFYTNRVLKDYKHSDLRHVDWVKSYLNIWSELQAYIKEHHTTGLTWSKTGP-VASTVSAF |
| .YV.EM.DAATFYTNRVLK.YKH.DL..VDWV.SY.NIWSE.QA..KEHHTTGLTWS.TGP..A.T.SA. | |
| Retrocopy | TYVNEMCDAATFYTNRVLKGYKH-DLHRVDWVRSYVNIWSEFQA*VKEHHTTGLTWSRTGP<AAPT-SAL |
| Parental | SVLSSGPGLPPPPPPL-PPPGP-PPLFENEGKKEESSPSRSALFAQLNQGEA---ITKGLRHVTDDQKTY |
| SVLS.GP.L.P.PP.L.PP.GP.PPLFENE..KE.SSPS.SALFA..NQGEA....T..L.H.......Y | |
| Retrocopy | SVLS*GPDLSPHPPHL<PP-GP<PPLFENEDTKEKSSPSCSALFARFNQGEA*PCRTRALAHH**PEGLY |
| Parental | KNPSLRAQGGQTQSPTKSHTPSPTSPKSYPSQKHAPVLELEGKKWRVEYQEDRNDLVISETELKQVAYIF |
| KNPSL.A.GGQ..SP..S..PSPTSPK..PSQ.HAPV..LEG.KWR.EYQEDRNDLVISETELKQVAYIF | |
| Retrocopy | KNPSLQAHGGQSTSPHRSYIPSPTSPKPHPSQQHAPVFKLEGSKWRAEYQEDRNDLVISETELKQVAYIF |
| Parental | KCEKSTIQIKGKVNSIIIDNCKKLGLVFDNVVGIVEVINSQDIQIQVMGRVPTISINKTEGCHIYLS |
| .CEKS....KGKVNSI...NCKK.GLVFD..VGI.E..NS..IQIQVMGRVP.IS.NKTEGCH.YL. | |
| Retrocopy | NCEKSALPMKGKVNSIPVENCKKFGLVFDSLVGIGEMTNSKYIQIQVMGRVPAISTNKTEGCHTYLT |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .14 RPM | 4 .47 RPM |
| bodymap2_adrenal | 0 .04 RPM | 20 .43 RPM |
| bodymap2_brain | 0 .00 RPM | 176 .81 RPM |
| bodymap2_breast | 0 .00 RPM | 78 .66 RPM |
| bodymap2_colon | 0 .00 RPM | 85 .04 RPM |
| bodymap2_heart | 0 .00 RPM | 116 .05 RPM |
| bodymap2_kidney | 0 .00 RPM | 22 .33 RPM |
| bodymap2_liver | 0 .00 RPM | 2 .77 RPM |
| bodymap2_lung | 0 .00 RPM | 11 .13 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 15 .53 RPM |
| bodymap2_ovary | 0 .08 RPM | 16 .93 RPM |
| bodymap2_prostate | 0 .00 RPM | 73 .25 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 871 .85 RPM |
| bodymap2_testis | 0 .00 RPM | 34 .11 RPM |
| bodymap2_thyroid | 0 .00 RPM | 94 .83 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 0 .00 RPM |
RNA Polymerase II activity near the 5' end of retro_hsap_1451 was not detected
No EST(s) were mapped for retro_hsap_1451 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
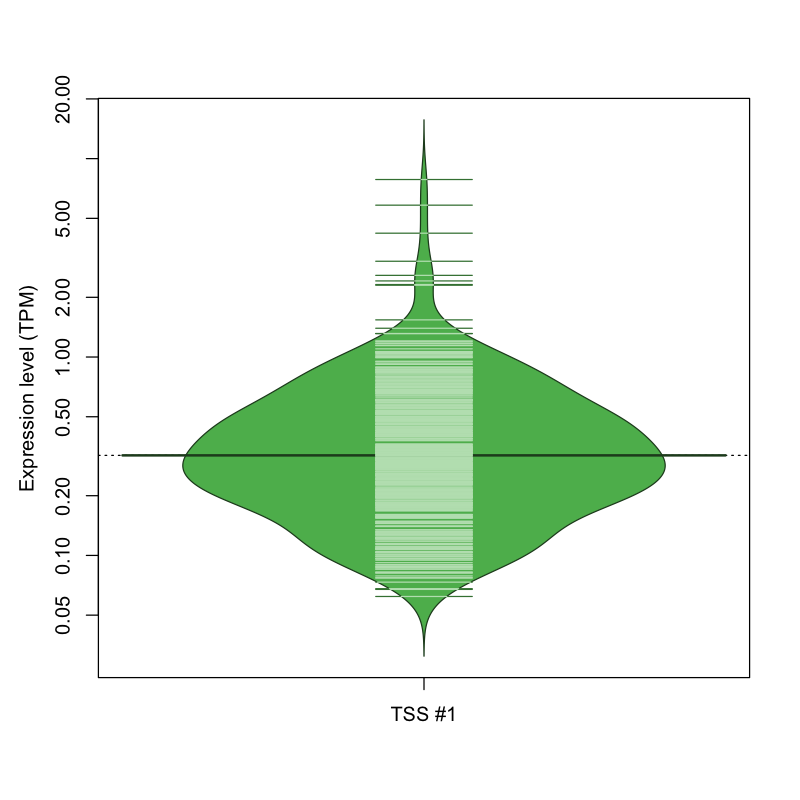
| TSS #1 | TSS_41219 | 1394 libraries | 408 libraries | 25 libraries | 2 libraries | 0 libraries |
The graphical summary, for retro_hsap_1451 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_1451 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_1451 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_987 |
| Macaca mulatta | retro_mmul_2271 |
Parental genes homology:
Parental genes homology involve 6 parental genes, and 7 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Canis familiaris | ENSCAFG00000009977 | 1 retrocopy | |
| Cavia porcellus | ENSCPOG00000008839 | 1 retrocopy | |
| Homo sapiens | ENSG00000112186 | 1 retrocopy |
retro_hsap_1451 ,
|
| Homo sapiens | ENSG00000131236 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000001990 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000017750 | 1 retrocopy |
Expression level across human populations :
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Undefined variable $populLEGEND in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3353
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .00 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .00 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .00 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .00 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .00 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .00 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .00 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .00 RPM |
Fatal error: Uncaught Error: Call to undefined function mysql_query() in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php:3390 Stack trace: #0 /home/rosikiewicz/RetrogeneDB2/BETA/retrogene.php(1158): include() #1 {main} thrown in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3390