RetrogeneDB ID: | retro_hsap_1699 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 16:69790440..69791840(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000260290 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | NONO | ||
| Ensembl ID: | ENSG00000147140 | ||
| Aliases: | NONO, NMT55, NRB54, P54, P54NRB | ||
| Description: | non-POU domain containing, octamer-binding [Source:HGNC Symbol;Acc:7871] |

Retrocopy-Parental alignment summary:
>retro_hsap_1699
ATGCAGAGTAATAAAACTTTTAACTTGGAGAAGCAAAACCATACTCCAAGAAAGCATCATCAACATCACCACCAGCAGCA
ACAGCTGCAGCCGCCACCACCGCCAATACCTGCAAATGGGCAACAGGCCAGCAGCCAAAATGAAGGCTTTGACTATTTGG
CCTGAAGAATTTTAGAAAACCAGGAGAGAAGACCTTCACCCAACGAAGCCGTCTTTTTGTGGGAAATCTTCCTCCCGACA
TCACTGAGGAAGAAATGAGGAAACTATTTGAGAAATATGGAAAGGCAGGCGAAGTCTTCATTCATAAGGATTAAGGATTT
GGCTTTATCCGCTTGGAAACACGAACCCTAGCGGAGATTGCGAAAGTGGAGCTGGACAATATGCCACTCTGTGGAAAGCA
GCTGCGTGTGCGCTTTTCCTGCCATAGTGCATCCCTTACAGTTCGAAACCTTCCTCAGTATGTGTCCAACGAAGTGCTGG
AAGAAGCCTTTTCTGTGTTTGGCCAGGTAGAGAGGGCTGTAGTCATTGTGGATGATCGAGGAAGGCCCTCAGGAAAAGGC
ATTGTTGAGTTCTCAGGGAAGCCAGCTGCTGGGAAAGCTCTGGACAGACGCAGTGAAGGCTCCTTCCTGCTAACCACATT
TCCTCGTCCTGTGACTATGGAGCCCATGGACCAGTTAGATGATGAAGGGGGACTTCCAGAGAAGCTGGTTATAAAAAACC
AGCAATTTCACAAGGAACGAGAGCAACCACCCAGATTTGCACAGCCTGGCTCCTTTGAGTATGAATATGCCATGCGCTGG
AAGGCACTCATTGAGATGGAGAAGCAGCAGCAGGACCAAGTGGACCGCAACATCAAAGAGGCTCGTGAGAAGCCAGAGAT
GGAGATGGAGGCTGCACGCCATAAGCACCAGGTCATGCTAATGAGACAGAATTTGATGAGGCGCCAAGAAGAACTTCGGA
GGATGGAAGAGCTGCACAACCAAGAGGTGCAAAAACGAAAGCAACTGGAGCTCAGGCAGGAGGAAGAGCGCAGGCGCCCT
GAAGAAGAGATGCGGCGGCAGCAAGAAGAAATGATGCAGCCACAGCAGGAAGGATTCAAGGGAACCTTCCCTGATGTGAG
AGAGCAGGAGATTTGGATGGGTCAGATGGCTATGGGAGGTGCTATGGGCATAAACAGCAGAGGTGCCTTGACCCCTGCTC
CTGTGCCAGCTGGTACCCCAGCTCCTCCAGGACCTGCCACTATGATGCCGGATGGAACTTTGGGATTGACCCCACCAACA
ACTGAACGCTTTGGTCAGGCTGCTACAATGGAAGGAATTGGGGCAATTGGTGAAACTCCTCCTGCATTCAACCATGCAGC
TCCTGGAGCTGAATTTGCTCCAAACAAACGTCGCCGATAC
ATGCAGAGTAATAAAACTTTTAACTTGGAGAAGCAAAACCATACTCCAAGAAAGCATCATCAACATCACCACCAGCAGCA
ACAGCTGCAGCCGCCACCACCGCCAATACCTGCAAATGGGCAACAGGCCAGCAGCCAAAATGAAGGCTTTGACTATTTGG
CCTGAAGAATTTTAGAAAACCAGGAGAGAAGACCTTCACCCAACGAAGCCGTCTTTTTGTGGGAAATCTTCCTCCCGACA
TCACTGAGGAAGAAATGAGGAAACTATTTGAGAAATATGGAAAGGCAGGCGAAGTCTTCATTCATAAGGATTAAGGATTT
GGCTTTATCCGCTTGGAAACACGAACCCTAGCGGAGATTGCGAAAGTGGAGCTGGACAATATGCCACTCTGTGGAAAGCA
GCTGCGTGTGCGCTTTTCCTGCCATAGTGCATCCCTTACAGTTCGAAACCTTCCTCAGTATGTGTCCAACGAAGTGCTGG
AAGAAGCCTTTTCTGTGTTTGGCCAGGTAGAGAGGGCTGTAGTCATTGTGGATGATCGAGGAAGGCCCTCAGGAAAAGGC
ATTGTTGAGTTCTCAGGGAAGCCAGCTGCTGGGAAAGCTCTGGACAGACGCAGTGAAGGCTCCTTCCTGCTAACCACATT
TCCTCGTCCTGTGACTATGGAGCCCATGGACCAGTTAGATGATGAAGGGGGACTTCCAGAGAAGCTGGTTATAAAAAACC
AGCAATTTCACAAGGAACGAGAGCAACCACCCAGATTTGCACAGCCTGGCTCCTTTGAGTATGAATATGCCATGCGCTGG
AAGGCACTCATTGAGATGGAGAAGCAGCAGCAGGACCAAGTGGACCGCAACATCAAAGAGGCTCGTGAGAAGCCAGAGAT
GGAGATGGAGGCTGCACGCCATAAGCACCAGGTCATGCTAATGAGACAGAATTTGATGAGGCGCCAAGAAGAACTTCGGA
GGATGGAAGAGCTGCACAACCAAGAGGTGCAAAAACGAAAGCAACTGGAGCTCAGGCAGGAGGAAGAGCGCAGGCGCCCT
GAAGAAGAGATGCGGCGGCAGCAAGAAGAAATGATGCAGCCACAGCAGGAAGGATTCAAGGGAACCTTCCCTGATGTGAG
AGAGCAGGAGATTTGGATGGGTCAGATGGCTATGGGAGGTGCTATGGGCATAAACAGCAGAGGTGCCTTGACCCCTGCTC
CTGTGCCAGCTGGTACCCCAGCTCCTCCAGGACCTGCCACTATGATGCCGGATGGAACTTTGGGATTGACCCCACCAACA
ACTGAACGCTTTGGTCAGGCTGCTACAATGGAAGGAATTGGGGCAATTGGTGAAACTCCTCCTGCATTCAACCATGCAGC
TCCTGGAGCTGAATTTGCTCCAAACAAACGTCGCCGATAC
ORF - retro_hsap_1699 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 90.06 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MQSNKTFNLEKQNHTPRKHHQHHHQQQHHQQQQQQPPPPPIPANGQQASSQNEG-LTI-DLKNFRKPGEK |
| MQSNKTFNLEKQNHTPRKHHQHHHQQQ....Q.Q.P.PPPIPANGQQASSQNEG.LTI..LKNFRKPGEK | |
| Retrocopy | MQSNKTFNLEKQNHTPRKHHQHHHQQQ----QLQPP-PPPIPANGQQASSQNEG>LTI>GLKNFRKPGEK |
| Parental | TFTQRSRLFVGNLPPDITEEEMRKLFEKYGKAGEVFIHKDKGFGFIRLETRTLAEIAKVELDNMPLRGKQ |
| TFTQRSRLFVGNLPPDITEEEMRKLFEKYGKAGEVFIHKD.GFGFIRLETRTLAEIAKVELDNMPL.GKQ | |
| Retrocopy | TFTQRSRLFVGNLPPDITEEEMRKLFEKYGKAGEVFIHKD*GFGFIRLETRTLAEIAKVELDNMPLCGKQ |
| Parental | LRVRFACHSASLTVRNLPQYVSNELLEEAFSVFGQVERAVVIVDDRGRPSGKGIVEFSGKPAARKALDRC |
| LRVRF.CHSASLTVRNLPQYVSNE.LEEAFSVFGQVERAVVIVDDRGRPSGKGIVEFSGKPAA.KALDR. | |
| Retrocopy | LRVRFSCHSASLTVRNLPQYVSNEVLEEAFSVFGQVERAVVIVDDRGRPSGKGIVEFSGKPAAGKALDRR |
| Parental | SEGSFLLTTFPRPVTVEPMDQLDDEEGLPEKLVIKNQQFHKEREQPPRFAQPGSFEYEYAMRWKALIEME |
| SEGSFLLTTFPRPVT.EPMDQLDDE.GLPEKLVIKNQQFHKEREQPPRFAQPGSFEYEYAMRWKALIEME | |
| Retrocopy | SEGSFLLTTFPRPVTMEPMDQLDDEGGLPEKLVIKNQQFHKEREQPPRFAQPGSFEYEYAMRWKALIEME |
| Parental | KQQQDQVDRNIKEAREKLEMEMEAARHEHQVMLMRQDLMRRQEELRRMEELHNQEVQKRKQLELRQEEER |
| KQQQDQVDRNIKEAREK.EMEMEAARH.HQVMLMRQ.LMRRQEELRRMEELHNQEVQKRKQLELRQEEER | |
| Retrocopy | KQQQDQVDRNIKEAREKPEMEMEAARHKHQVMLMRQNLMRRQEELRRMEELHNQEVQKRKQLELRQEEER |
| Parental | RRREEEMRRQQEEMMRRQQEGFKGTFPDAREQEIRMGQMAMGGAMGINNRGAMPPAPVPAGTPAPPGPAT |
| RR...................FKGTFPD.REQEI.MGQMAMGGAMGIN.RGA..PAPVPAGTPAPPGPAT | |
| Retrocopy | RRPXXXXXXXXXXXXXXXXXXFKGTFPDVREQEIWMGQMAMGGAMGINSRGALTPAPVPAGTPAPPGPAT |
| Parental | MMPDGTLGLTPPTTERFGQAATMEGIGAIGGTPPAFNRAAPGAEFAPNKRRRY |
| MMPDGTLGLTPPTTERFGQAATMEGIGAIG.TPPAFN.AAPGAEFAPNKRRRY | |
| Retrocopy | MMPDGTLGLTPPTTERFGQAATMEGIGAIGETPPAFNHAAPGAEFAPNKRRRY |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .16 RPM | 219 .73 RPM |
| bodymap2_adrenal | 0 .06 RPM | 212 .51 RPM |
| bodymap2_brain | 0 .07 RPM | 80 .73 RPM |
| bodymap2_breast | 0 .06 RPM | 175 .66 RPM |
| bodymap2_colon | 0 .02 RPM | 176 .52 RPM |
| bodymap2_heart | 0 .00 RPM | 70 .17 RPM |
| bodymap2_kidney | 0 .08 RPM | 155 .74 RPM |
| bodymap2_liver | 0 .06 RPM | 96 .87 RPM |
| bodymap2_lung | 0 .11 RPM | 137 .20 RPM |
| bodymap2_lymph_node | 0 .02 RPM | 211 .65 RPM |
| bodymap2_ovary | 0 .15 RPM | 373 .01 RPM |
| bodymap2_prostate | 0 .12 RPM | 228 .49 RPM |
| bodymap2_skeletal_muscle | 0 .11 RPM | 86 .59 RPM |
| bodymap2_testis | 0 .08 RPM | 197 .66 RPM |
| bodymap2_thyroid | 0 .06 RPM | 208 .71 RPM |
| bodymap2_white_blood_cells | 0 .37 RPM | 271 .48 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_1699 was not detected
No EST(s) were mapped for retro_hsap_1699 retrocopy.
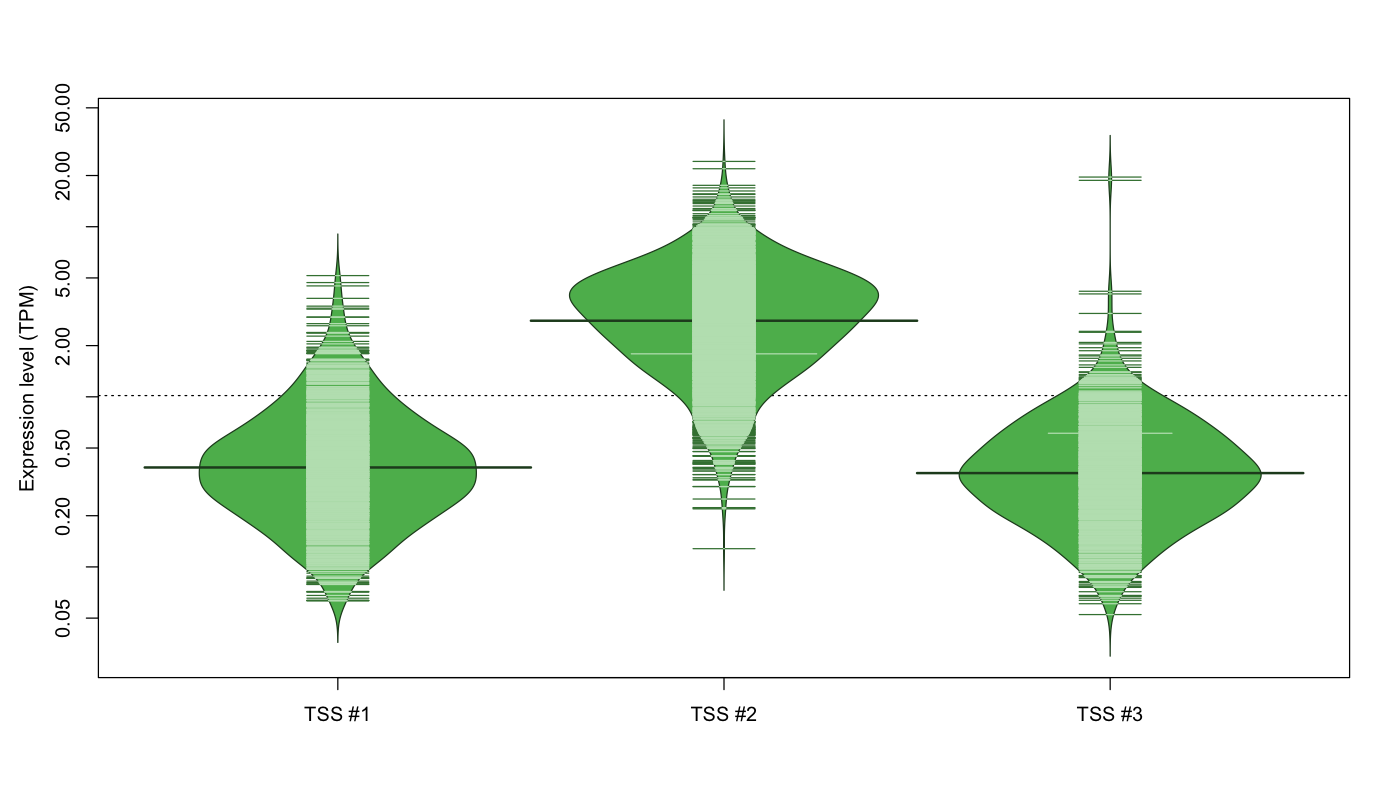
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_53485 | 1016 libraries | 721 libraries | 91 libraries | 1 library | 0 libraries |
| TSS #2 | TSS_53486 | 187 libraries | 150 libraries | 1139 libraries | 320 libraries | 33 libraries |
| TSS #3 | TSS_53487 | 997 libraries | 779 libraries | 51 libraries | 0 libraries | 2 libraries |
The graphical summary, for retro_hsap_1699 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_1699 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_1699 has 3 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_1150 |
| Gorilla gorilla | retro_ggor_1273 |
| Pongo abelii | retro_pabe_1398 |
Parental genes homology:
Parental genes homology involve 21 parental genes, and 31 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .17 RPM |
| CEU_NA11843 | 0 .11 RPM |
| CEU_NA11930 | 0 .49 RPM |
| CEU_NA12004 | 0 .04 RPM |
| CEU_NA12400 | 0 .25 RPM |
| CEU_NA12751 | 0 .15 RPM |
| CEU_NA12760 | 0 .63 RPM |
| CEU_NA12827 | 0 .14 RPM |
| CEU_NA12872 | 0 .35 RPM |
| CEU_NA12873 | 0 .13 RPM |
| FIN_HG00183 | 0 .16 RPM |
| FIN_HG00277 | 0 .41 RPM |
| FIN_HG00315 | 0 .19 RPM |
| FIN_HG00321 | 0 .21 RPM |
| FIN_HG00328 | 0 .54 RPM |
| FIN_HG00338 | 0 .28 RPM |
| FIN_HG00349 | 0 .26 RPM |
| FIN_HG00375 | 0 .29 RPM |
| FIN_HG00377 | 0 .25 RPM |
| FIN_HG00378 | 0 .34 RPM |
| GBR_HG00099 | 0 .35 RPM |
| GBR_HG00111 | 0 .17 RPM |
| GBR_HG00114 | 0 .13 RPM |
| GBR_HG00119 | 0 .22 RPM |
| GBR_HG00131 | 0 .17 RPM |
| GBR_HG00133 | 0 .31 RPM |
| GBR_HG00134 | 0 .20 RPM |
| GBR_HG00137 | 0 .16 RPM |
| GBR_HG00142 | 0 .22 RPM |
| GBR_HG00143 | 0 .26 RPM |
| TSI_NA20512 | 0 .23 RPM |
| TSI_NA20513 | 0 .29 RPM |
| TSI_NA20518 | 0 .44 RPM |
| TSI_NA20532 | 0 .17 RPM |
| TSI_NA20538 | 0 .46 RPM |
| TSI_NA20756 | 0 .58 RPM |
| TSI_NA20765 | 0 .29 RPM |
| TSI_NA20771 | 0 .23 RPM |
| TSI_NA20786 | 0 .31 RPM |
| TSI_NA20798 | 0 .25 RPM |
| YRI_NA18870 | 0 .58 RPM |
| YRI_NA18907 | 0 .10 RPM |
| YRI_NA18916 | 0 .21 RPM |
| YRI_NA19093 | 0 .10 RPM |
| YRI_NA19099 | 0 .48 RPM |
| YRI_NA19114 | 0 .26 RPM |
| YRI_NA19118 | 0 .19 RPM |
| YRI_NA19213 | 0 .45 RPM |
| YRI_NA19214 | 0 .17 RPM |
| YRI_NA19223 | 0 .32 RPM |