RetrogeneDB ID: | retro_hsap_2422 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 20:30135306..30135849(+) | ||
| Located in intron of: | ENSG00000101294 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000101898 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | MCTS1 | ||
| Ensembl ID: | ENSG00000232119 | ||
| Aliases: | None | ||
| Description: | malignant T cell amplified sequence 1 [Source:HGNC Symbol;Acc:23357] |

Retrocopy-Parental alignment summary:
>retro_hsap_2422
ATGTTCAAGAAGTTTGATGAAAAGGAAAGTGTGTCCAACTGCATCCAGTTGAAAACGTCAGTTATTAAGGGCATTAAGAG
CCAACTGGTAGAGCAATTTCCAGGTATTGAACCATGGCTTAATCAAATCATGCCTAAGAAAGATCCTGTCAAAATAGTCC
GATGCCACGAACATACAGAAATCCTTACCGTAAGTGGGGAATTATTGTTTTTTAGACAAAGAAAGGGGCCTTTTTGTCCA
ACTCTAAGGTTGCTTCACAAATACCCTTTTATCCTGCCACACCAGCAGGTTGATAAAGGAGCTATCAAATTTGTACTCAG
TGGCGCAAATATTATGTGTCCAGGTTTAACTTCTCCTGGAGCTAAGCTGTACCCTGCTGCAGTAGATACGATTGTAGCAG
TCACAGCGGAAGGAAAACAGCATGCTCTGTGTGTTGGGGTCATGAAGATGTCTGCAGAAGATATTGAGAAAGTCAACAAA
GGAATTGGCATTGAAAATATCCATTATTTAAATGATGGGCTGTGGCACATGAAGACATATAAA
ATGTTCAAGAAGTTTGATGAAAAGGAAAGTGTGTCCAACTGCATCCAGTTGAAAACGTCAGTTATTAAGGGCATTAAGAG
CCAACTGGTAGAGCAATTTCCAGGTATTGAACCATGGCTTAATCAAATCATGCCTAAGAAAGATCCTGTCAAAATAGTCC
GATGCCACGAACATACAGAAATCCTTACCGTAAGTGGGGAATTATTGTTTTTTAGACAAAGAAAGGGGCCTTTTTGTCCA
ACTCTAAGGTTGCTTCACAAATACCCTTTTATCCTGCCACACCAGCAGGTTGATAAAGGAGCTATCAAATTTGTACTCAG
TGGCGCAAATATTATGTGTCCAGGTTTAACTTCTCCTGGAGCTAAGCTGTACCCTGCTGCAGTAGATACGATTGTAGCAG
TCACAGCGGAAGGAAAACAGCATGCTCTGTGTGTTGGGGTCATGAAGATGTCTGCAGAAGATATTGAGAAAGTCAACAAA
GGAATTGGCATTGAAAATATCCATTATTTAAATGATGGGCTGTGGCACATGAAGACATATAAA
ORF - retro_hsap_2422 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 95.03 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MFKKFDEKENVSNCIQLKTSVIKGIKNQLIEQFPGIEPWLNQIMPKKDPVKIVRCHEHIEILTVNGELLF |
| MFKKFDEKE.VSNCIQLKTSVIKGIK.QL.EQFPGIEPWLNQIMPKKDPVKIVRCHEH.EILTV.GELLF | |
| Retrocopy | MFKKFDEKESVSNCIQLKTSVIKGIKSQLVEQFPGIEPWLNQIMPKKDPVKIVRCHEHTEILTVSGELLF |
| Parental | FRQREGPFYPTLRLLHKYPFILPHQQVDKGAIKFVLSGANIMCPGLTSPGAKLYPAAVDTIVAIMAEGKQ |
| FRQR.GPF.PTLRLLHKYPFILPHQQVDKGAIKFVLSGANIMCPGLTSPGAKLYPAAVDTIVA..AEGKQ | |
| Retrocopy | FRQRKGPFCPTLRLLHKYPFILPHQQVDKGAIKFVLSGANIMCPGLTSPGAKLYPAAVDTIVAVTAEGKQ |
| Parental | HALCVGVMKMSAEDIEKVNKGIGIENIHYLNDGLWHMKTYK |
| HALCVGVMKMSAEDIEKVNKGIGIENIHYLNDGLWHMKTYK | |
| Retrocopy | HALCVGVMKMSAEDIEKVNKGIGIENIHYLNDGLWHMKTYK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 1 .03 RPM | 16 .26 RPM |
| bodymap2_adrenal | 2 .01 RPM | 18 .38 RPM |
| bodymap2_brain | 1 .31 RPM | 18 .21 RPM |
| bodymap2_breast | 0 .72 RPM | 23 .29 RPM |
| bodymap2_colon | 1 .10 RPM | 22 .74 RPM |
| bodymap2_heart | 0 .70 RPM | 16 .01 RPM |
| bodymap2_kidney | 0 .60 RPM | 26 .20 RPM |
| bodymap2_liver | 0 .34 RPM | 29 .21 RPM |
| bodymap2_lung | 1 .02 RPM | 15 .51 RPM |
| bodymap2_lymph_node | 1 .48 RPM | 22 .92 RPM |
| bodymap2_ovary | 3 .08 RPM | 15 .02 RPM |
| bodymap2_prostate | 0 .94 RPM | 22 .38 RPM |
| bodymap2_skeletal_muscle | 0 .53 RPM | 12 .66 RPM |
| bodymap2_testis | 1 .73 RPM | 20 .96 RPM |
| bodymap2_thyroid | 1 .75 RPM | 14 .84 RPM |
| bodymap2_white_blood_cells | 0 .35 RPM | 26 .01 RPM |
RNA Polymerase II actvity may be related with retro_hsap_2422 in 48 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CGN | POLR2A | 20:30135101..30135364 |
| ENCFF002CHO | POLR2A | 20:30134756..30135447 |
| ENCFF002CIH | POLR2A | 20:30134806..30135382 |
| ENCFF002CIO | POLR2A | 20:30134880..30135425 |
| ENCFF002CJE | POLR2A | 20:30134821..30135491 |
| ENCFF002CJZ | POLR2A | 20:30134816..30135463 |
| ENCFF002CKX | POLR2A | 20:30134874..30135409 |
| ENCFF002CLM | POLR2A | 20:30134966..30135506 |
| ENCFF002CMI | POLR2A | 20:30134806..30135433 |
| ENCFF002COJ | POLR2A | 20:30134992..30135428 |
| ENCFF002CPG | POLR2A | 20:30135130..30135359 |
| ENCFF002CPH | POLR2A | 20:30134867..30135257 |
| ENCFF002CQA | POLR2A | 20:30135172..30135368 |
| ENCFF002CQC | POLR2A | 20:30135093..30135376 |
| ENCFF002CQE | POLR2A | 20:30134995..30135355 |
| ENCFF002CQG | POLR2A | 20:30135098..30135337 |
| ENCFF002CQI | POLR2A | 20:30135086..30135378 |
| ENCFF002CQK | POLR2A | 20:30134957..30135390 |
| ENCFF002CQM | POLR2A | 20:30135045..30135380 |
| ENCFF002CQO | POLR2A | 20:30134942..30135370 |
| ENCFF002CRK | POLR2A | 20:30134999..30135523 |
| ENCFF002CRO | POLR2A | 20:30134996..30135416 |
| ENCFF002CSY | POLR2A | 20:30134805..30135511 |
| ENCFF002CUP | POLR2A | 20:30135136..30135270 |
| ENCFF002CUQ | POLR2A | 20:30134798..30135414 |
| ENCFF002CVF | POLR2A | 20:30135029..30135445 |
| ENCFF002CVJ | POLR2A | 20:30134864..30135165 |
| ENCFF002CVJ | POLR2A | 20:30135163..30135399 |
| ENCFF002CXM | POLR2A | 20:30134815..30135305 |
| ENCFF002CXN | POLR2A | 20:30134796..30135292 |
| ENCFF002CXN | POLR2A | 20:30134997..30135493 |
| ENCFF002CXO | POLR2A | 20:30135038..30135462 |
| ENCFF002CXP | POLR2A | 20:30135104..30135233 |
| ENCFF002CXR | POLR2A | 20:30135094..30135264 |
| ENCFF002CZC | POLR2A | 20:30134846..30135454 |
| ENCFF002CZD | POLR2A | 20:30134899..30135401 |
| ENCFF002CZQ | POLR2A | 20:30135065..30135345 |
| ENCFF002CZW | POLR2A | 20:30134838..30135494 |
| ENCFF002CZY | POLR2A | 20:30134911..30135392 |
| ENCFF002DAH | POLR2A | 20:30135056..30135340 |
| ENCFF002DAK | POLR2A | 20:30135009..30135509 |
| ENCFF002DAS | POLR2A | 20:30134973..30135449 |
| ENCFF002DAV | POLR2A | 20:30135094..30135263 |
| ENCFF002DAY | POLR2A | 20:30134917..30135553 |
| ENCFF002DBB | POLR2A | 20:30135131..30135331 |
| ENCFF002DBE | POLR2A | 20:30135193..30135317 |
| ENCFF002DBO | POLR2A | 20:30135064..30135318 |
| ENCFF002DBP | POLR2A | 20:30135029..30135359 |
| ENCFF002DBQ | POLR2A | 20:30134936..30135306 |
| ENCFF002DBT | POLR2A | 20:30134950..30135320 |
No EST(s) were mapped for retro_hsap_2422 retrocopy.
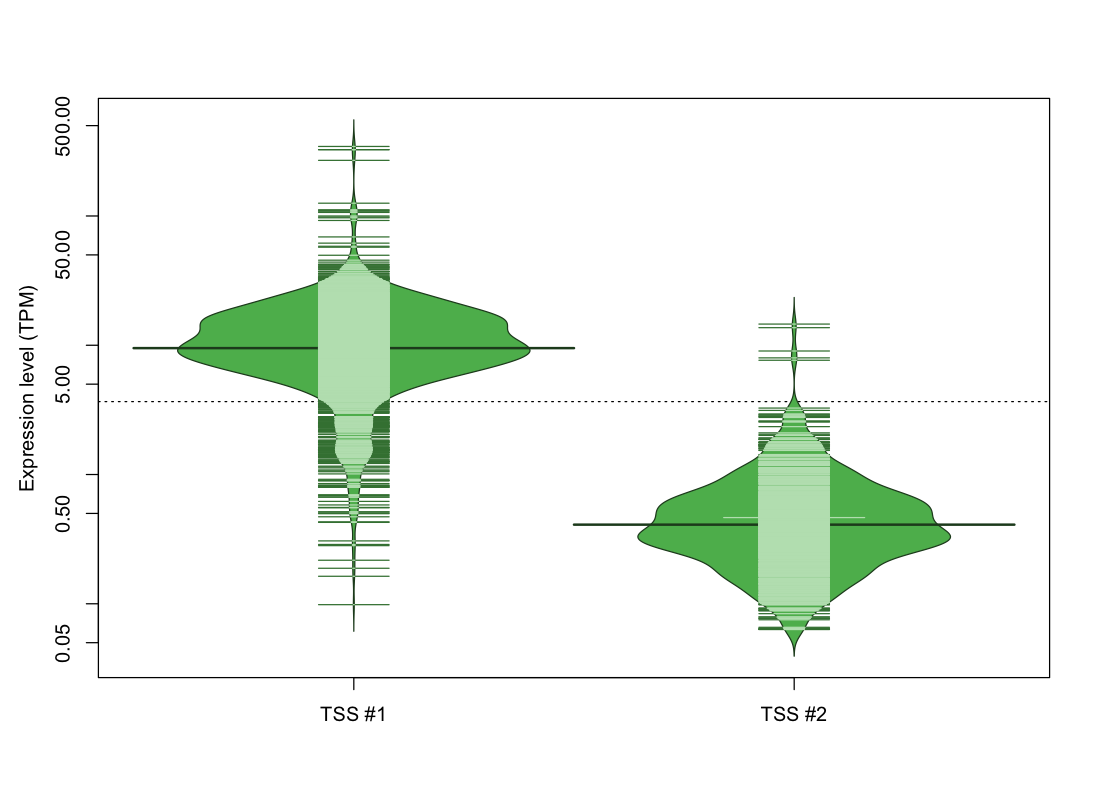
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_99479 | 104 libraries | 33 libraries | 210 libraries | 610 libraries | 872 libraries |
| TSS #2 | TSS_99480 | 1079 libraries | 659 libraries | 86 libraries | 3 libraries | 2 libraries |
The graphical summary, for retro_hsap_2422 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | CCTGCAGGGGACTCCACCGGAGC (23 nt long) |
| Reverse primer: | TTCATCTGTGGTAAGAGCACCC (22 nt long) |
| Anneling temperature: | 63 °C |
| (Expected) product size: | 785 |
| Electrophoresis gel image: | 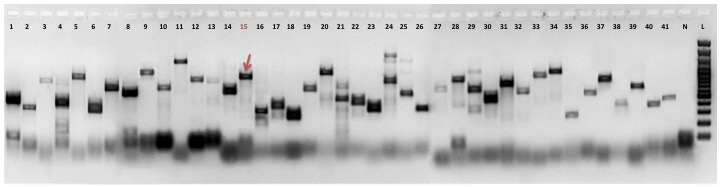 |
| Additional comment: | Red arrow indicates PCR product of retrogene in pooled cDNA from 16 human tissues (Clontech). Red number of lane indicates particular retrogene; L - GeneRuler 100 bp DNA Ladder (Thermo Fisher Scientific); N - No template control (water instead of cDNA) |
Retrocopy orthology:
Retrocopy retro_hsap_2422 has 6 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 16 parental genes, and 23 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Cavia porcellus | ENSCPOG00000005502 | 1 retrocopy | |
| Erinaceus europaeus | ENSEEUG00000007258 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000015462 | 1 retrocopy | |
| Homo sapiens | ENSG00000232119 | 1 retrocopy |
retro_hsap_2422 ,
|
| Gorilla gorilla | ENSGGOG00000009420 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000015101 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000010354 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000000355 | 3 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000014315 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000022241 | 1 retrocopy | |
| Pteropus vampyrus | ENSPVAG00000013249 | 3 retrocopies | |
| Rattus norvegicus | ENSRNOG00000002563 | 2 retrocopies | |
| Ictidomys tridecemlineatus | ENSSTOG00000011496 | 2 retrocopies | |
| Tupaia belangeri | ENSTBEG00000010666 | 2 retrocopies | |
| Tarsius syrichta | ENSTSYG00000001440 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000007208 | 1 retrocopy |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 2 .50 RPM |
| CEU_NA11843 | 2 .13 RPM |
| CEU_NA11930 | 3 .72 RPM |
| CEU_NA12004 | 3 .87 RPM |
| CEU_NA12400 | 2 .00 RPM |
| CEU_NA12751 | 3 .38 RPM |
| CEU_NA12760 | 3 .19 RPM |
| CEU_NA12827 | 3 .44 RPM |
| CEU_NA12872 | 3 .84 RPM |
| CEU_NA12873 | 4 .02 RPM |
| FIN_HG00183 | 3 .38 RPM |
| FIN_HG00277 | 2 .40 RPM |
| FIN_HG00315 | 2 .79 RPM |
| FIN_HG00321 | 3 .17 RPM |
| FIN_HG00328 | 3 .03 RPM |
| FIN_HG00338 | 3 .11 RPM |
| FIN_HG00349 | 3 .98 RPM |
| FIN_HG00375 | 3 .77 RPM |
| FIN_HG00377 | 3 .45 RPM |
| FIN_HG00378 | 3 .55 RPM |
| GBR_HG00099 | 3 .60 RPM |
| GBR_HG00111 | 2 .94 RPM |
| GBR_HG00114 | 2 .76 RPM |
| GBR_HG00119 | 3 .07 RPM |
| GBR_HG00131 | 3 .13 RPM |
| GBR_HG00133 | 3 .28 RPM |
| GBR_HG00134 | 3 .10 RPM |
| GBR_HG00137 | 2 .39 RPM |
| GBR_HG00142 | 4 .03 RPM |
| GBR_HG00143 | 3 .67 RPM |
| TSI_NA20512 | 2 .29 RPM |
| TSI_NA20513 | 4 .04 RPM |
| TSI_NA20518 | 3 .08 RPM |
| TSI_NA20532 | 3 .98 RPM |
| TSI_NA20538 | 4 .70 RPM |
| TSI_NA20756 | 2 .82 RPM |
| TSI_NA20765 | 3 .71 RPM |
| TSI_NA20771 | 3 .74 RPM |
| TSI_NA20786 | 3 .71 RPM |
| TSI_NA20798 | 3 .62 RPM |
| YRI_NA18870 | 3 .72 RPM |
| YRI_NA18907 | 3 .33 RPM |
| YRI_NA18916 | 3 .47 RPM |
| YRI_NA19093 | 4 .45 RPM |
| YRI_NA19099 | 3 .70 RPM |
| YRI_NA19114 | 3 .41 RPM |
| YRI_NA19118 | 4 .18 RPM |
| YRI_NA19213 | 2 .54 RPM |
| YRI_NA19214 | 3 .39 RPM |
| YRI_NA19223 | 3 .70 RPM |