RetrogeneDB ID: | retro_hsap_3087 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 4:120314453..120316112(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000249244 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | GK | ||
| Ensembl ID: | ENSG00000198814 | ||
| Aliases: | GK, GK1, GKD | ||
| Description: | glycerol kinase [Source:HGNC Symbol;Acc:4289] |

Retrocopy-Parental alignment summary:
>retro_hsap_3087
ATGGCAGCCTCAAAAAAGGCAGTTTTGGGGCCATTGGTGGGGGCAGTGGACCAGGGCACCAGTTCCACGCGCTTTTTGGT
TTTCAATTCAAGAACAGCTGAACTACTTAGTCATCATCAAGTGGAAATAAAACAAGAGTTCCCAAGAGAAGGATGGGTGG
AACAGGACCCTAAGGAAATTCTACATTCTGTCTATGAGTGTATAGCGAAAACATGTGAGAAACTTGGACAGCTCAATATT
GGTATTTCCAACATAAAAGCTATTGGTGTCAGCAACCAGAGGGAAACCACTGTAGTCTGGGACAAGATAACTGGAGAGCC
TCTCTACAATGCTGTGGTGTGGCTTGAACTAAGAACACAGTCTACCGTTGAGAGTCTTAGTAAGAGAATTCCAGGAAATA
ATAACTTTGTCAAGTCCAAGACAGGCCTTCCACTTAACACTTACTTCAGTGCAGTGAAACTTCGCTGGCTCCTTGACAAT
GTGAGAAAAGTTCAAAAGGCCGTTGAAGAAAAACGAGCTCTTTTTGGGACTATTGATTCATGGCTTATTTGGAGTTTGAC
AGGAGGCGTCAATGGAGGTGTCCACTGTACAGATGTAACAAATGCAAGTAGGACTATGCTTTTCAACATTCATTCTTTGG
AATGGGATAAACAACTCTGTGAATTTTTTGGAATTCCAACGGAAATTCTTCCACATGTTCGGAGTTCTTCTGAGATCTAT
GGCCTAATGAAAGCTGGGGCCTTGGAAGGTGTGCCAATATCTGGGTGTTCAGGGGACCAGTCTGCTGCACTGGTGGGACA
AATGTGTTTCCAGATTGGACAAGCCAAAAATACGTATGGAACAGGATATTTCTTACTATGTAATACATGCCATAAGTGTG
TATTTTCTGATCATGGCCTTCTCACCACAATGGCTTACAAACTTGGCAGAGACAAACCGGTATATTATGCTTTGGAAGGT
TCTGTAGCTATAGCTGGTGCTGTTATTCGCTGGCTAAGAGACAATCTTGGAATTATAAAGACCTCAGAAGAAATTGAAAA
ACTTGCTAAAGAAGTAGGTACTTCTTATGGCAGCTACTTCGTCCCAGCATTTTCGGGGTTATATGCACCTTATTGGGAGC
CCAGCGCAAGAGGGATAATCTGTGGACTCACTCAGTTTACCAATAAATGCCATATTGCTTTTGCTGCATTAGAAGCTGTT
TGTTTCCATATTCGAGAGATTTTGGATGCCATGAATTGAGACTGTGGAATTCCACTCAGTCATTTGCAGGTTGATGGAGG
AAGGACCAGCAACAAAATTCTTATGCAGCTACAAGCAGACATTCTGTATATTCCAGTAGTGAAGCCCTTGATGCCCGAAA
CCACTGCATTGGGTGCTGCCATGGCGGCAGGGGCTGCAGAAGGAGTCGTCGTATGGAGTCTTGAACCTGAGGATTTCTCC
GCCGTCACAATGGAGCGGTTTGAACCTCAGATTAATGCTGAGGAAAGTGAAATTCGTTATTCTACATGGAAGAAAGCTGT
GATGAAGTCAATGGGTTGGGTTACAACTCAATCTCCAGAAAGTGGTGACCCTAGTGTCTTCTGTAGTCTGCCCTTGGGCT
TTTTTATAGTGAGTAGCATGGCAATGTTAATCAGAGCAAGGTACATCTCAGGTATTCCA
ATGGCAGCCTCAAAAAAGGCAGTTTTGGGGCCATTGGTGGGGGCAGTGGACCAGGGCACCAGTTCCACGCGCTTTTTGGT
TTTCAATTCAAGAACAGCTGAACTACTTAGTCATCATCAAGTGGAAATAAAACAAGAGTTCCCAAGAGAAGGATGGGTGG
AACAGGACCCTAAGGAAATTCTACATTCTGTCTATGAGTGTATAGCGAAAACATGTGAGAAACTTGGACAGCTCAATATT
GGTATTTCCAACATAAAAGCTATTGGTGTCAGCAACCAGAGGGAAACCACTGTAGTCTGGGACAAGATAACTGGAGAGCC
TCTCTACAATGCTGTGGTGTGGCTTGAACTAAGAACACAGTCTACCGTTGAGAGTCTTAGTAAGAGAATTCCAGGAAATA
ATAACTTTGTCAAGTCCAAGACAGGCCTTCCACTTAACACTTACTTCAGTGCAGTGAAACTTCGCTGGCTCCTTGACAAT
GTGAGAAAAGTTCAAAAGGCCGTTGAAGAAAAACGAGCTCTTTTTGGGACTATTGATTCATGGCTTATTTGGAGTTTGAC
AGGAGGCGTCAATGGAGGTGTCCACTGTACAGATGTAACAAATGCAAGTAGGACTATGCTTTTCAACATTCATTCTTTGG
AATGGGATAAACAACTCTGTGAATTTTTTGGAATTCCAACGGAAATTCTTCCACATGTTCGGAGTTCTTCTGAGATCTAT
GGCCTAATGAAAGCTGGGGCCTTGGAAGGTGTGCCAATATCTGGGTGTTCAGGGGACCAGTCTGCTGCACTGGTGGGACA
AATGTGTTTCCAGATTGGACAAGCCAAAAATACGTATGGAACAGGATATTTCTTACTATGTAATACATGCCATAAGTGTG
TATTTTCTGATCATGGCCTTCTCACCACAATGGCTTACAAACTTGGCAGAGACAAACCGGTATATTATGCTTTGGAAGGT
TCTGTAGCTATAGCTGGTGCTGTTATTCGCTGGCTAAGAGACAATCTTGGAATTATAAAGACCTCAGAAGAAATTGAAAA
ACTTGCTAAAGAAGTAGGTACTTCTTATGGCAGCTACTTCGTCCCAGCATTTTCGGGGTTATATGCACCTTATTGGGAGC
CCAGCGCAAGAGGGATAATCTGTGGACTCACTCAGTTTACCAATAAATGCCATATTGCTTTTGCTGCATTAGAAGCTGTT
TGTTTCCATATTCGAGAGATTTTGGATGCCATGAATTGAGACTGTGGAATTCCACTCAGTCATTTGCAGGTTGATGGAGG
AAGGACCAGCAACAAAATTCTTATGCAGCTACAAGCAGACATTCTGTATATTCCAGTAGTGAAGCCCTTGATGCCCGAAA
CCACTGCATTGGGTGCTGCCATGGCGGCAGGGGCTGCAGAAGGAGTCGTCGTATGGAGTCTTGAACCTGAGGATTTCTCC
GCCGTCACAATGGAGCGGTTTGAACCTCAGATTAATGCTGAGGAAAGTGAAATTCGTTATTCTACATGGAAGAAAGCTGT
GATGAAGTCAATGGGTTGGGTTACAACTCAATCTCCAGAAAGTGGTGACCCTAGTGTCTTCTGTAGTCTGCCCTTGGGCT
TTTTTATAGTGAGTAGCATGGCAATGTTAATCAGAGCAAGGTACATCTCAGGTATTCCA
ORF - retro_hsap_3087 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 96.02 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAASKKAVLGPLVGAVDQGTSSTRFLVFNSKTAELLSHHQVEIKQEFPREGWVEQDPKEILHSVYECIEK |
| MAASKKAVLGPLVGAVDQGTSSTRFLVFNS.TAELLSHHQVEIKQEFPREGWVEQDPKEILHSVYECI.K | |
| Retrocopy | MAASKKAVLGPLVGAVDQGTSSTRFLVFNSRTAELLSHHQVEIKQEFPREGWVEQDPKEILHSVYECIAK |
| Parental | TCEKLGQLNIDISNIKAIGVSNQRETTVVWDKITGEPLYNAVVWLDLRTQSTVESLSKRIPGNNNFVKSK |
| TCEKLGQLNI.ISNIKAIGVSNQRETTVVWDKITGEPLYNAVVWL.LRTQSTVESLSKRIPGNNNFVKSK | |
| Retrocopy | TCEKLGQLNIGISNIKAIGVSNQRETTVVWDKITGEPLYNAVVWLELRTQSTVESLSKRIPGNNNFVKSK |
| Parental | TGLPLSTYFSAVKLRWLLDNVRKVQKAVEEKRALFGTIDSWLIWSLTGGVNGGVHCTDVTNASRTMLFNI |
| TGLPL.TYFSAVKLRWLLDNVRKVQKAVEEKRALFGTIDSWLIWSLTGGVNGGVHCTDVTNASRTMLFNI | |
| Retrocopy | TGLPLNTYFSAVKLRWLLDNVRKVQKAVEEKRALFGTIDSWLIWSLTGGVNGGVHCTDVTNASRTMLFNI |
| Parental | HSLEWDKQLCEFFGIPMEILPNVRSSSEIYGLMKAGALEGVPISGCLGDQSAALVGQMCFQIGQAKNTYG |
| HSLEWDKQLCEFFGIP.EILP.VRSSSEIYGLMKAGALEGVPISGC.GDQSAALVGQMCFQIGQAKNTYG | |
| Retrocopy | HSLEWDKQLCEFFGIPTEILPHVRSSSEIYGLMKAGALEGVPISGCSGDQSAALVGQMCFQIGQAKNTYG |
| Parental | TGCFLLCNTGHKCVFSDHGLLTTVAYKLGRDKPVYYALEGSVAIAGAVIRWLRDNLGIIKTSEEIEKLAK |
| TG.FLLCNT.HKCVFSDHGLLTT.AYKLGRDKPVYYALEGSVAIAGAVIRWLRDNLGIIKTSEEIEKLAK | |
| Retrocopy | TGYFLLCNTCHKCVFSDHGLLTTMAYKLGRDKPVYYALEGSVAIAGAVIRWLRDNLGIIKTSEEIEKLAK |
| Parental | EVGTSYGCYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFQTREILDAMNRDCGIPLS |
| EVGTSYG.YFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCF..REILDAMN.DCGIPLS | |
| Retrocopy | EVGTSYGSYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFHIREILDAMN*DCGIPLS |
| Parental | HLQVDGGMTSNKILMQLQADILYIPVVKPSMPETTALGAAMAAGAAEGVGVWSLEPEDLSAVTMERFEPQ |
| HLQVDGG.TSNKILMQLQADILYIPVVKP.MPETTALGAAMAAGAAEGV.VWSLEPED.SAVTMERFEPQ | |
| Retrocopy | HLQVDGGRTSNKILMQLQADILYIPVVKPLMPETTALGAAMAAGAAEGVVVWSLEPEDFSAVTMERFEPQ |
| Parental | INAEESEIRYSTWKKAVMKSMGWVTTQSPESGDPSIFCSLPLGFFIVSSMVMLIGARYISGIP |
| INAEESEIRYSTWKKAVMKSMGWVTTQSPESGDPS.FCSLPLGFFIVSSM.MLI.ARYISGIP | |
| Retrocopy | INAEESEIRYSTWKKAVMKSMGWVTTQSPESGDPSVFCSLPLGFFIVSSMAMLIRARYISGIP |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 4 .37 RPM |
| bodymap2_adrenal | 0 .04 RPM | 4 .46 RPM |
| bodymap2_brain | 0 .23 RPM | 9 .47 RPM |
| bodymap2_breast | 0 .21 RPM | 3 .07 RPM |
| bodymap2_colon | 0 .21 RPM | 2 .03 RPM |
| bodymap2_heart | 0 .33 RPM | 5 .48 RPM |
| bodymap2_kidney | 0 .06 RPM | 16 .75 RPM |
| bodymap2_liver | 0 .09 RPM | 45 .33 RPM |
| bodymap2_lung | 0 .00 RPM | 18 .74 RPM |
| bodymap2_lymph_node | 0 .16 RPM | 8 .90 RPM |
| bodymap2_ovary | 0 .40 RPM | 6 .25 RPM |
| bodymap2_prostate | 0 .00 RPM | 3 .41 RPM |
| bodymap2_skeletal_muscle | 0 .29 RPM | 0 .45 RPM |
| bodymap2_testis | 2 .88 RPM | 7 .47 RPM |
| bodymap2_thyroid | 0 .26 RPM | 6 .55 RPM |
| bodymap2_white_blood_cells | 0 .20 RPM | 25 .63 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_3087 was not detected
No EST(s) were mapped for retro_hsap_3087 retrocopy.
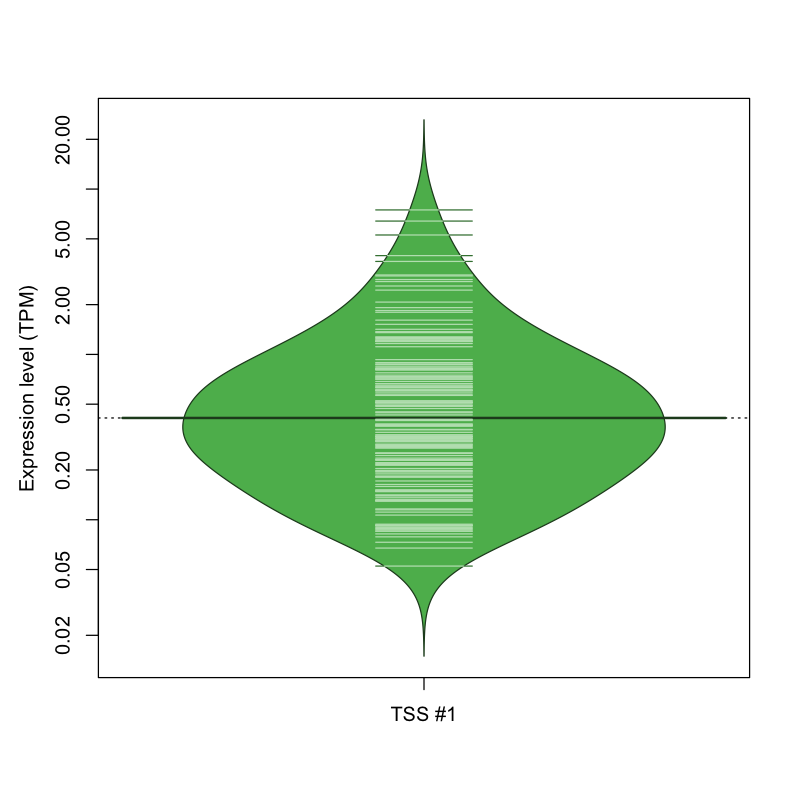
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_139350 | 1666 libraries | 133 libraries | 27 libraries | 3 libraries | 0 libraries |
The graphical summary, for retro_hsap_3087 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_3087 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_3087 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 14 parental genes, and 25 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000007633 | 3 retrocopies | |
| Cavia porcellus | ENSCPOG00000003261 | 2 retrocopies | |
| Echinops telfairi | ENSETEG00000008279 | 2 retrocopies | |
| Felis catus | ENSFCAG00000004264 | 1 retrocopy | |
| Homo sapiens | ENSG00000198814 | 4 retrocopies | |
| Loxodonta africana | ENSLAFG00000007632 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000002417 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000025059 | 2 retrocopies | |
| Nomascus leucogenys | ENSNLEG00000010102 | 2 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000013023 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000034116 | 2 retrocopies | |
| Sarcophilus harrisii | ENSSHAG00000006470 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000008732 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000008249 | 1 retrocopy |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .11 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .07 RPM |
| CEU_NA12004 | 0 .04 RPM |
| CEU_NA12400 | 0 .18 RPM |
| CEU_NA12751 | 0 .02 RPM |
| CEU_NA12760 | 0 .13 RPM |
| CEU_NA12827 | 0 .09 RPM |
| CEU_NA12872 | 0 .05 RPM |
| CEU_NA12873 | 0 .32 RPM |
| FIN_HG00183 | 0 .19 RPM |
| FIN_HG00277 | 0 .33 RPM |
| FIN_HG00315 | 0 .17 RPM |
| FIN_HG00321 | 0 .18 RPM |
| FIN_HG00328 | 0 .45 RPM |
| FIN_HG00338 | 0 .06 RPM |
| FIN_HG00349 | 0 .03 RPM |
| FIN_HG00375 | 0 .07 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .21 RPM |
| GBR_HG00099 | 0 .06 RPM |
| GBR_HG00111 | 0 .02 RPM |
| GBR_HG00114 | 0 .13 RPM |
| GBR_HG00119 | 0 .31 RPM |
| GBR_HG00131 | 0 .06 RPM |
| GBR_HG00133 | 0 .05 RPM |
| GBR_HG00134 | 0 .11 RPM |
| GBR_HG00137 | 0 .05 RPM |
| GBR_HG00142 | 0 .19 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .08 RPM |
| TSI_NA20513 | 0 .07 RPM |
| TSI_NA20518 | 0 .06 RPM |
| TSI_NA20532 | 0 .20 RPM |
| TSI_NA20538 | 0 .37 RPM |
| TSI_NA20756 | 0 .06 RPM |
| TSI_NA20765 | 0 .08 RPM |
| TSI_NA20771 | 0 .11 RPM |
| TSI_NA20786 | 0 .08 RPM |
| TSI_NA20798 | 0 .03 RPM |
| YRI_NA18870 | 0 .03 RPM |
| YRI_NA18907 | 0 .10 RPM |
| YRI_NA18916 | 0 .08 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .21 RPM |
| YRI_NA19114 | 0 .13 RPM |
| YRI_NA19118 | 0 .12 RPM |
| YRI_NA19213 | 0 .17 RPM |
| YRI_NA19214 | 0 .07 RPM |
| YRI_NA19223 | 0 .14 RPM |