RetrogeneDB ID: | retro_mmus_97 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 18:52693721..52695371(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000053624 | |
| Aliases: | Gykl1, AI449806, Gk-rs1 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Gyk | ||
| Ensembl ID: | ENSMUSG00000025059 | ||
| Aliases: | Gyk, D930012N15Rik, GK | ||
| Description: | glycerol kinase [Source:MGI Symbol;Acc:MGI:106594] |

Retrocopy-Parental alignment summary:
>retro_mmus_97
ATGGCAGAGGCTCCAAAAGCGGTTTTGGGGCCGTTGGTAGGGGCAGTGGACCAAGGTACCAGCTCAACACGTTTTTTGGT
TTTCAATCCCCAAACAGCTGAATTACTTTGTCATCATCAAGTGGAAATAGCCCAGGAGTTCCCAAAAGAAGGATGGGTAG
AGCAAGACCCAAAGGCAATCCTGCAGTCTGTGTACGAGTGTATAGAGAAAGCATGTGAGAAACTCGGACAGCAGAGCATT
GATATTTCCAACATAAAAGCTATCGGTGTCACCAACCAGCGGGAAACTACGATAGTCTGGGACAAGTTCACTGGAGAACC
TCTCTACAATGCTGTGGTGTGGCTTGACCTAAGGACCCAGTCTACTGTGGAGAATCTCAGCAAAAGTATTTCAGTAAGCA
ATAACTTTGTCAAGAACAAGACAGGCCTTCCAATCAGCACTTATTTTAGTGCCGTGAAACTTCACTGGCTCATTGAAAAT
GTGAGAAAAGTGCAAAAGGCTATTGAAGATGGCAGAGCTATTTTTGGGACGGTTGATTCGTGGCTTATCTGGTGTATGAC
TGGAGGTATCAACGGAGGTGTCCACTGTACAGATGTAAGTAATGCCAGCAGGACCATGCTTTTCAACATTCACTCTTTGC
AATGGGATGAAGAGCTCTGCGACTTTTTTGGAATTCCAATGACCATCCTTCCCCGAATCCGGAGTTCTTCTGAGATCTAT
GGCCTAGTGAAAAGTGGCGTGTTGGAAGGTGTGCCAATATCTGGATGTCTGGGAGACCAGTCTGCTGCTTTGGTGGGACA
ACTGTGTCTTCAGGACGGACAAGCTAAAAGCACATATGGAACAGGCTGTTTCTTACTATGTAACACAGGCCAGAAGTGTG
TAAACTCTGAACATGGCCTTCTGACCACAGTGGCTTACCAGCTTGGCAGACAGGAGCCTGTCTATTATGCCCTGGAAGGT
TCCGTAGCCATAGCCGGTGCTGTTGTTAGCTGGATAAAAAACAATCTTCAAATTATTCAGTCTTCATCGGAAATTGAAAA
ACTTGCTGAAGTAGCAGGGACTTCTTATGGTTGTTACTTCGTCCCAGCCTTTTCAGGCTTATACGCTCCATATTGGGACC
CAAGTGCGAGAGGGATCATCTGCGGGCTCACCCAGTTCACCAATAAGTTCCATATTGCCTTTGCTGCGTTAGAAGCTGTT
TGCTTCCAAACGCGGGAGATTTTGGATGCCATGAACAGTGATTGTGGAATCCCGCTGACGCATTTGCAGGTAGATGGAGG
AATGACCGCAAACAGAGTTCTCATGCAACTGCAGGCGGACATTCTGTGTATTCCGGTGATGAAACCTTTCATGCCAGAAA
CAACTGCACTGGGAGTCGCAATGGCAGCGGGAGCGGCAGAAGGAGTTAGCGTGTGGAGTCTGGATCCTAAGGATTTGTCA
AGTGTCCAGATGGAGAAGTTTGAACCCCAGATCAATGTCGAGGAAAGTGAAGGTCGTTACGCCACATGGAAGAAAGCTGT
GCTGAAGTCAATGGGTTGGGTTTTGACTCAGTCTCCTGATGGTCGCGACCCTAGTATTTTCAGCAGCCTGACGCTGGGCT
TTTATGTAGTGAGTAGTATGGTGATATTAATTGGAGCAAGGTACATGTAA
ATGGCAGAGGCTCCAAAAGCGGTTTTGGGGCCGTTGGTAGGGGCAGTGGACCAAGGTACCAGCTCAACACGTTTTTTGGT
TTTCAATCCCCAAACAGCTGAATTACTTTGTCATCATCAAGTGGAAATAGCCCAGGAGTTCCCAAAAGAAGGATGGGTAG
AGCAAGACCCAAAGGCAATCCTGCAGTCTGTGTACGAGTGTATAGAGAAAGCATGTGAGAAACTCGGACAGCAGAGCATT
GATATTTCCAACATAAAAGCTATCGGTGTCACCAACCAGCGGGAAACTACGATAGTCTGGGACAAGTTCACTGGAGAACC
TCTCTACAATGCTGTGGTGTGGCTTGACCTAAGGACCCAGTCTACTGTGGAGAATCTCAGCAAAAGTATTTCAGTAAGCA
ATAACTTTGTCAAGAACAAGACAGGCCTTCCAATCAGCACTTATTTTAGTGCCGTGAAACTTCACTGGCTCATTGAAAAT
GTGAGAAAAGTGCAAAAGGCTATTGAAGATGGCAGAGCTATTTTTGGGACGGTTGATTCGTGGCTTATCTGGTGTATGAC
TGGAGGTATCAACGGAGGTGTCCACTGTACAGATGTAAGTAATGCCAGCAGGACCATGCTTTTCAACATTCACTCTTTGC
AATGGGATGAAGAGCTCTGCGACTTTTTTGGAATTCCAATGACCATCCTTCCCCGAATCCGGAGTTCTTCTGAGATCTAT
GGCCTAGTGAAAAGTGGCGTGTTGGAAGGTGTGCCAATATCTGGATGTCTGGGAGACCAGTCTGCTGCTTTGGTGGGACA
ACTGTGTCTTCAGGACGGACAAGCTAAAAGCACATATGGAACAGGCTGTTTCTTACTATGTAACACAGGCCAGAAGTGTG
TAAACTCTGAACATGGCCTTCTGACCACAGTGGCTTACCAGCTTGGCAGACAGGAGCCTGTCTATTATGCCCTGGAAGGT
TCCGTAGCCATAGCCGGTGCTGTTGTTAGCTGGATAAAAAACAATCTTCAAATTATTCAGTCTTCATCGGAAATTGAAAA
ACTTGCTGAAGTAGCAGGGACTTCTTATGGTTGTTACTTCGTCCCAGCCTTTTCAGGCTTATACGCTCCATATTGGGACC
CAAGTGCGAGAGGGATCATCTGCGGGCTCACCCAGTTCACCAATAAGTTCCATATTGCCTTTGCTGCGTTAGAAGCTGTT
TGCTTCCAAACGCGGGAGATTTTGGATGCCATGAACAGTGATTGTGGAATCCCGCTGACGCATTTGCAGGTAGATGGAGG
AATGACCGCAAACAGAGTTCTCATGCAACTGCAGGCGGACATTCTGTGTATTCCGGTGATGAAACCTTTCATGCCAGAAA
CAACTGCACTGGGAGTCGCAATGGCAGCGGGAGCGGCAGAAGGAGTTAGCGTGTGGAGTCTGGATCCTAAGGATTTGTCA
AGTGTCCAGATGGAGAAGTTTGAACCCCAGATCAATGTCGAGGAAAGTGAAGGTCGTTACGCCACATGGAAGAAAGCTGT
GCTGAAGTCAATGGGTTGGGTTTTGACTCAGTCTCCTGATGGTCGCGACCCTAGTATTTTCAGCAGCCTGACGCTGGGCT
TTTATGTAGTGAGTAGTATGGTGATATTAATTGGAGCAAGGTACATGTAA
ORF - retro_mmus_97 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 82.88 % |
| Parental protein coverage: | 99.28 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAAAKKAVLGPLVGAVDQGTSSTRFLVFNSKTAELLSHHQVEIKQEFPREGWVEQDPKEILQSVYECIEK |
| MA.A.KAVLGPLVGAVDQGTSSTRFLVFN..TAELL.HHQVEI.QEFP.EGWVEQDPK.ILQSVYECIEK | |
| Retrocopy | MAEAPKAVLGPLVGAVDQGTSSTRFLVFNPQTAELLCHHQVEIAQEFPKEGWVEQDPKAILQSVYECIEK |
| Parental | TCEKLGQLNIDISNIKAIGVSNQRETTVVWDKVTGEPLYNAVVWLDLRTQSTVENLSKRIPGNNNFVKSK |
| .CEKLGQ..IDISNIKAIGV.NQRETT.VWDK.TGEPLYNAVVWLDLRTQSTVENLSK.I...NNFVK.K | |
| Retrocopy | ACEKLGQQSIDISNIKAIGVTNQRETTIVWDKFTGEPLYNAVVWLDLRTQSTVENLSKSISVSNNFVKNK |
| Parental | TGLPLSTYFSAVKLRWLLDNVKKVQEAVEENRALFGTIDSWLIWSLTGGIHGGVHCTDVTNASRTMLFNI |
| TGLP.STYFSAVKL.WL..NV.KVQ.A.E..RA.FGT.DSWLIW..TGGI.GGVHCTDV.NASRTMLFNI | |
| Retrocopy | TGLPISTYFSAVKLHWLIENVRKVQKAIEDGRAIFGTVDSWLIWCMTGGINGGVHCTDVSNASRTMLFNI |
| Parental | HSLEWDKELCEFFGIPMEILPNVRSSSEIYGLMKAGALEGVPISGCLGDQSAALVGQMCFQDGQAKNTYG |
| HSL.WD.ELC.FFGIPM.ILP..RSSSEIYGL.K.G.LEGVPISGCLGDQSAALVGQ.C.QDGQAK.TYG | |
| Retrocopy | HSLQWDEELCDFFGIPMTILPRIRSSSEIYGLVKSGVLEGVPISGCLGDQSAALVGQLCLQDGQAKSTYG |
| Parental | TGCFLLCNTGHKCVFSEHGLLTTVAYKLGRDKPVYYALEGSVAIAGAVIRWLRDNLGIIKSSEEIEKLAK |
| TGCFLLCNTG.KCV.SEHGLLTTVAY.LGR..PVYYALEGSVAIAGAV..W...NL.II.SS.EIEKLA. | |
| Retrocopy | TGCFLLCNTGQKCVNSEHGLLTTVAYQLGRQEPVYYALEGSVAIAGAVVSWIKNNLQIIQSSSEIEKLAE |
| Parental | EVGTSYGCYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFQTREILDAMNRDCGIPLS |
| ..GTSYGCYFVPAFSGLYAPYW.PSARGIICGLTQFTNK.HIAFAALEAVCFQTREILDAMN.DCGIPL. | |
| Retrocopy | VAGTSYGCYFVPAFSGLYAPYWDPSARGIICGLTQFTNKFHIAFAALEAVCFQTREILDAMNSDCGIPLT |
| Parental | HLQVDGGMTSNKILMQLQADILYIPVVKPSMPETTALGAAMAAGAAEGVGVWSLEPEDLSAVTMERFEPQ |
| HLQVDGGMT.N..LMQLQADIL.IPV.KP.MPETTALG.AMAAGAAEGV.VWSL.P.DLS.V.ME.FEPQ | |
| Retrocopy | HLQVDGGMTANRVLMQLQADILCIPVMKPFMPETTALGVAMAAGAAEGVSVWSLDPKDLSSVQMEKFEPQ |
| Parental | INAEESEIRYSTWKKAVMKSIGWVTTQSPESGDPSIFCSLPLGFFIVSSMVMLIGARYI |
| IN.EESE.RY.TWKKAV.KS.GWV.TQSP...DPSIF.SL.LGF..VSSMV.LIGARY. | |
| Retrocopy | INVEESEGRYATWKKAVLKSMGWVLTQSPDGRDPSIFSSLTLGFYVVSSMVILIGARYM |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 18 .83 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 15 .68 RPM |
| SRP007412_heart | 0 .00 RPM | 23 .60 RPM |
| SRP007412_kidney | 0 .00 RPM | 331 .61 RPM |
| SRP007412_liver | 0 .05 RPM | 75 .92 RPM |
| SRP007412_testis | 412 .29 RPM | 1 .56 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_97 was not detected
19 EST(s) were mapped to retro_mmus_97 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA063880 | 52693720 | 52694216 | 98 | 485 | 9 | 473 |
| AA064451 | 52693715 | 52693838 | 99.2 | 122 | 1 | 121 |
| AA065397 | 52693724 | 52693826 | 99.1 | 101 | 1 | 100 |
| AA107211 | 52693727 | 52694319 | 99.2 | 585 | 5 | 579 |
| AA107620 | 52693734 | 52694217 | 97.5 | 468 | 9 | 457 |
| AA492930 | 52694279 | 52694398 | 97.5 | 116 | 3 | 113 |
| AI647419 | 52694339 | 52694672 | 99.4 | 329 | 2 | 326 |
| BB572574 | 52693689 | 52693965 | 96.8 | 267 | 9 | 258 |
| BB614799 | 52693706 | 52694352 | 98.5 | 633 | 9 | 621 |
| BF013306 | 52693858 | 52694191 | 100 | 333 | 0 | 333 |
| BF149591 | 52693736 | 52694318 | 99.2 | 572 | 4 | 565 |
| BF149654 | 52693766 | 52694281 | 100 | 515 | 0 | 515 |
| BG100740 | 52694785 | 52695079 | 100 | 292 | 0 | 290 |
| BG101398 | 52693964 | 52694342 | 98.2 | 371 | 7 | 364 |
| BU962784 | 52693689 | 52694460 | 98.9 | 770 | 0 | 766 |
| BY094838 | 52693690 | 52694081 | 99.3 | 388 | 3 | 385 |
| BY353966 | 52694111 | 52694370 | 97.7 | 252 | 4 | 244 |
| CA463392 | 52693875 | 52694517 | 100 | 635 | 0 | 628 |
| CF105239 | 52693714 | 52694290 | 99.7 | 574 | 2 | 572 |
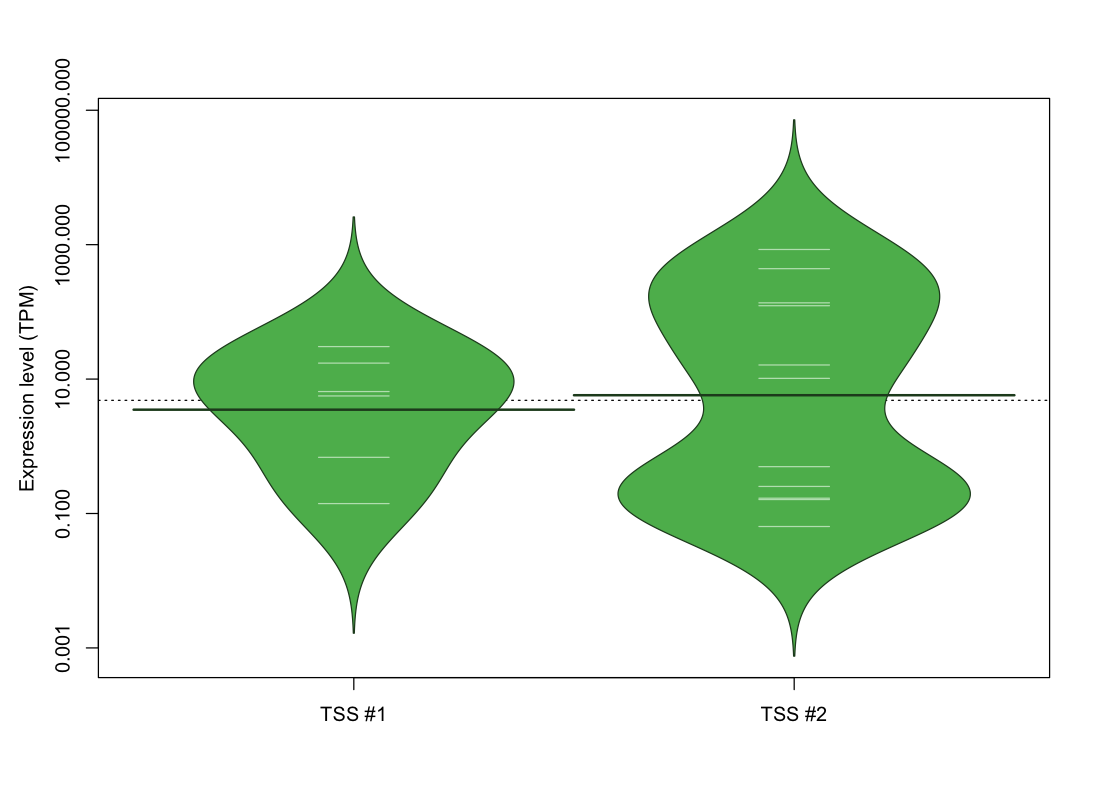
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_59780 | 1066 libraries | 2 libraries | 0 libraries | 2 libraries | 2 libraries |
| TSS #2 | TSS_59781 | 1061 libraries | 5 libraries | 0 libraries | 0 libraries | 6 libraries |
The graphical summary, for retro_mmus_97 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_97 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_97 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_114 |
Parental genes homology:
Parental genes homology involve 14 parental genes, and 25 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000007633 | 3 retrocopies | |
| Cavia porcellus | ENSCPOG00000003261 | 2 retrocopies | |
| Echinops telfairi | ENSETEG00000008279 | 2 retrocopies | |
| Felis catus | ENSFCAG00000004264 | 1 retrocopy | |
| Homo sapiens | ENSG00000198814 | 4 retrocopies | |
| Loxodonta africana | ENSLAFG00000007632 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000002417 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000025059 | 2 retrocopies |
retro_mmus_119, retro_mmus_97 ,
|
| Nomascus leucogenys | ENSNLEG00000010102 | 2 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000013023 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000034116 | 2 retrocopies | |
| Sarcophilus harrisii | ENSSHAG00000006470 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000008732 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000008249 | 1 retrocopy |