RetrogeneDB ID: | retro_mmus_119 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 5:97455312..97456977(-) | ||
| Located in intron of: | ENSMUSG00000097516 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000050553 | |
| Aliases: | Gk2, GKTA, Gk-rs2 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Gyk | ||
| Ensembl ID: | ENSMUSG00000025059 | ||
| Aliases: | Gyk, D930012N15Rik, GK | ||
| Description: | glycerol kinase [Source:MGI Symbol;Acc:MGI:106594] |

Retrocopy-Parental alignment summary:
>retro_mmus_119
ATGGCAGCCTCGAAGCAAACCTCTGCGGGACCGTTGGTAGGAGCGGTGGTCCAGGGAACCAACTCCACTCGCTTTCTAGT
TTTCAATAGCAAAACAGCAGAACTAGTTTGCTCTCATCAGGTGGAACTGACACAAGAATACCCAAAAGAAGGATGGGTGG
AGCAAGACCCCAAGGAAATCCTTAAATCTGTCTATGAGTGCATAGCGAAAGCTTGTGAAAAACTTGCTGAAGTGAACATT
GATATTTCCAACATTAAAGCTATCGGCGTGAGTAACCAGCGGGAAACCACTGTGGTCTGGGACAAGTTCACGGGAGACCC
TCTGTACAATGCCGTGGTGTGGCTTGATCTCAGAACCCAGTCTACTGTGGAGACTCTCACTAAGAAAATCCCAGGAAATA
GCAACTTTGTCAAGTCTAAGACCGGCCTTCCTCTTAGCACTTACTTCAGTGCTGTGAAGCTGCGGTGGATGCTGGACAAC
CTAAGACCCATTCAAAAGGCTGTTGAGGAAGGCAGAGCCATGTTTGGAACCATCGACTCTTGGCTCATCTGGTGTATGAC
CGGAGGAGTCAATGGAGGCATCCATTGCACCGATGTAACAAATGCATGCAGAACGATGCTTTTTAACATCCACTCTTTGG
AATGGGATAAAGATCTCTGTGACTTTTTTGAAATTCCAATGAGCATCCTCCCAAATGTATGCAGTTCTTCTGAGATCTAT
GGCCTGATGACATCAGGGGCCTTGGAAGGCGTGCCAATATCTGGATGTTTGGGGGATCAGTCTGCTGCTTTAGTGGGACA
AATGTGTTTCCATGAAGGTCAAGCCAAAAACACATACGGAACAGGCTGTTTTTTACTATGTAATACAGGCCAGAAATGTG
TGTTTTCTGAACATGGCCTTCTGACCACATTGGCTTACAAACTAGGCAAAAATAAGCCAGTTTTTTATGCCTTAGAAGGT
TCTGTTGCTATAGCTGGTGCTGTTATTCGCTGGCTAAGAGACAATTTCGAAATTATAACAACCTCAGGGGAAGTTGAAAA
TCTTGCAAGAGAGGTAGGCACTTCCTATGGCTGCTACTTTGTCCCAGCCTTTTCGGGGCTGTATGCACCTTACTGGGAAC
CCAGTGCAAGAGGGATAATCTGTGGTCTCACTCAGTTCACCAACAAATGCCATATTGCCTTTGCTGCGTTAGAAGCTGTT
TGTTTCCAAACCCGAGAGATTTTGGATGCCATGAATCGCGACTGTGGAATTCCACTCAGTCATTTGCAGGTCGATGGAGG
AATGACCAACAACAGGATTCTTATGCAGCTACAGGCAGATATTCTGCACATTCCGGTAGTTAAGTCTGTCATGCCTGAAA
CGACGGCCCTGGGAGCTGCCATGGCAGCCGGAGCTGCAGAAGGGGTAAATGTTTGGAGTCTTGAACCTGAAGATTTGTCT
ACGATCTTGATGGAACGATATGAACCACAAATCCAAGCCACAGAGAGTGAAATTCGTTTTTCAACATGGAAGAGAGCAGT
GATGAAGTCAATGGGTTGGGTTACAGCAAAGGACCCTGAAAATGGCGATAACCCTGTCTTCTCTTGTCTGCCCTTGGGTT
TTTTTATAGTGAGTAGCATGACGTTATTAATTGGAGCAAGATGTGTCTCAACTGCTGAAGAGTAA
ATGGCAGCCTCGAAGCAAACCTCTGCGGGACCGTTGGTAGGAGCGGTGGTCCAGGGAACCAACTCCACTCGCTTTCTAGT
TTTCAATAGCAAAACAGCAGAACTAGTTTGCTCTCATCAGGTGGAACTGACACAAGAATACCCAAAAGAAGGATGGGTGG
AGCAAGACCCCAAGGAAATCCTTAAATCTGTCTATGAGTGCATAGCGAAAGCTTGTGAAAAACTTGCTGAAGTGAACATT
GATATTTCCAACATTAAAGCTATCGGCGTGAGTAACCAGCGGGAAACCACTGTGGTCTGGGACAAGTTCACGGGAGACCC
TCTGTACAATGCCGTGGTGTGGCTTGATCTCAGAACCCAGTCTACTGTGGAGACTCTCACTAAGAAAATCCCAGGAAATA
GCAACTTTGTCAAGTCTAAGACCGGCCTTCCTCTTAGCACTTACTTCAGTGCTGTGAAGCTGCGGTGGATGCTGGACAAC
CTAAGACCCATTCAAAAGGCTGTTGAGGAAGGCAGAGCCATGTTTGGAACCATCGACTCTTGGCTCATCTGGTGTATGAC
CGGAGGAGTCAATGGAGGCATCCATTGCACCGATGTAACAAATGCATGCAGAACGATGCTTTTTAACATCCACTCTTTGG
AATGGGATAAAGATCTCTGTGACTTTTTTGAAATTCCAATGAGCATCCTCCCAAATGTATGCAGTTCTTCTGAGATCTAT
GGCCTGATGACATCAGGGGCCTTGGAAGGCGTGCCAATATCTGGATGTTTGGGGGATCAGTCTGCTGCTTTAGTGGGACA
AATGTGTTTCCATGAAGGTCAAGCCAAAAACACATACGGAACAGGCTGTTTTTTACTATGTAATACAGGCCAGAAATGTG
TGTTTTCTGAACATGGCCTTCTGACCACATTGGCTTACAAACTAGGCAAAAATAAGCCAGTTTTTTATGCCTTAGAAGGT
TCTGTTGCTATAGCTGGTGCTGTTATTCGCTGGCTAAGAGACAATTTCGAAATTATAACAACCTCAGGGGAAGTTGAAAA
TCTTGCAAGAGAGGTAGGCACTTCCTATGGCTGCTACTTTGTCCCAGCCTTTTCGGGGCTGTATGCACCTTACTGGGAAC
CCAGTGCAAGAGGGATAATCTGTGGTCTCACTCAGTTCACCAACAAATGCCATATTGCCTTTGCTGCGTTAGAAGCTGTT
TGTTTCCAAACCCGAGAGATTTTGGATGCCATGAATCGCGACTGTGGAATTCCACTCAGTCATTTGCAGGTCGATGGAGG
AATGACCAACAACAGGATTCTTATGCAGCTACAGGCAGATATTCTGCACATTCCGGTAGTTAAGTCTGTCATGCCTGAAA
CGACGGCCCTGGGAGCTGCCATGGCAGCCGGAGCTGCAGAAGGGGTAAATGTTTGGAGTCTTGAACCTGAAGATTTGTCT
ACGATCTTGATGGAACGATATGAACCACAAATCCAAGCCACAGAGAGTGAAATTCGTTTTTCAACATGGAAGAGAGCAGT
GATGAAGTCAATGGGTTGGGTTACAGCAAAGGACCCTGAAAATGGCGATAACCCTGTCTTCTCTTGTCTGCCCTTGGGTT
TTTTTATAGTGAGTAGCATGACGTTATTAATTGGAGCAAGATGTGTCTCAACTGCTGAAGAGTAA
ORF - retro_mmus_119 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 83.64 % |
| Parental protein coverage: | 99.46 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MAAAKKAVLGPLVGAVDQGTSSTRFLVFNSKTAELLSHHQVEIKQEFPREGWVEQDPKEILQSVYECIEK |
| MAA.K....GPLVGAV.QGT.STRFLVFNSKTAEL...HQVE..QE.P.EGWVEQDPKEIL.SVYECI.K | |
| Retrocopy | MAASKQTSAGPLVGAVVQGTNSTRFLVFNSKTAELVCSHQVELTQEYPKEGWVEQDPKEILKSVYECIAK |
| Parental | TCEKLGQLNIDISNIKAIGVSNQRETTVVWDKVTGEPLYNAVVWLDLRTQSTVENLSKRIPGNNNFVKSK |
| .CEKL...NIDISNIKAIGVSNQRETTVVWDK.TG.PLYNAVVWLDLRTQSTVE.L.K.IPGN.NFVKSK | |
| Retrocopy | ACEKLAEVNIDISNIKAIGVSNQRETTVVWDKFTGDPLYNAVVWLDLRTQSTVETLTKKIPGNSNFVKSK |
| Parental | TGLPLSTYFSAVKLRWLLDNVKKVQEAVEENRALFGTIDSWLIWSLTGGIHGGVHCTDVTNASRTMLFNI |
| TGLPLSTYFSAVKLRW.LDN....Q.AVEE.RA.FGTIDSWLIW..TGG..GG.HCTDVTNA.RTMLFNI | |
| Retrocopy | TGLPLSTYFSAVKLRWMLDNLRPIQKAVEEGRAMFGTIDSWLIWCMTGGVNGGIHCTDVTNACRTMLFNI |
| Parental | HSLEWDKELCEFFGIPMEILPNVRSSSEIYGLMKAGALEGVPISGCLGDQSAALVGQMCFQDGQAKNTYG |
| HSLEWDK.LC.FF.IPM.ILPNV.SSSEIYGLM..GALEGVPISGCLGDQSAALVGQMCF..GQAKNTYG | |
| Retrocopy | HSLEWDKDLCDFFEIPMSILPNVCSSSEIYGLMTSGALEGVPISGCLGDQSAALVGQMCFHEGQAKNTYG |
| Parental | TGCFLLCNTGHKCVFSEHGLLTTVAYKLGRDKPVYYALEGSVAIAGAVIRWLRDNLGIIKSSEEIEKLAK |
| TGCFLLCNTG.KCVFSEHGLLTT.AYKLG..KPV.YALEGSVAIAGAVIRWLRDN..II..S.E.E.LA. | |
| Retrocopy | TGCFLLCNTGQKCVFSEHGLLTTLAYKLGKNKPVFYALEGSVAIAGAVIRWLRDNFEIITTSGEVENLAR |
| Parental | EVGTSYGCYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFQTREILDAMNRDCGIPLS |
| EVGTSYGCYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFQTREILDAMNRDCGIPLS | |
| Retrocopy | EVGTSYGCYFVPAFSGLYAPYWEPSARGIICGLTQFTNKCHIAFAALEAVCFQTREILDAMNRDCGIPLS |
| Parental | HLQVDGGMTSNKILMQLQADILYIPVVKPSMPETTALGAAMAAGAAEGVGVWSLEPEDLSAVTMERFEPQ |
| HLQVDGGMT.N.ILMQLQADIL.IPVVK..MPETTALGAAMAAGAAEGV.VWSLEPEDLS...MER.EPQ | |
| Retrocopy | HLQVDGGMTNNRILMQLQADILHIPVVKSVMPETTALGAAMAAGAAEGVNVWSLEPEDLSTILMERYEPQ |
| Parental | INAEESEIRYSTWKKAVMKSIGWVTTQSPESGDPSIFCSLPLGFFIVSSMVMLIGARYIS |
| I.A.ESEIR.STWK.AVMKS.GWVT...PE.GD...F..LPLGFFIVSSM..LIGAR..S | |
| Retrocopy | IQATESEIRFSTWKRAVMKSMGWVTAKDPENGDNPVFSCLPLGFFIVSSMTLLIGARCVS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .09 RPM | 18 .83 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 15 .68 RPM |
| SRP007412_heart | 0 .00 RPM | 23 .60 RPM |
| SRP007412_kidney | 0 .00 RPM | 331 .61 RPM |
| SRP007412_liver | 0 .11 RPM | 75 .92 RPM |
| SRP007412_testis | 401 .68 RPM | 1 .56 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_119 was not detected
No EST(s) were mapped for retro_mmus_119 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_116758 | 1061 libraries | 4 libraries | 1 library | 0 libraries | 6 libraries |
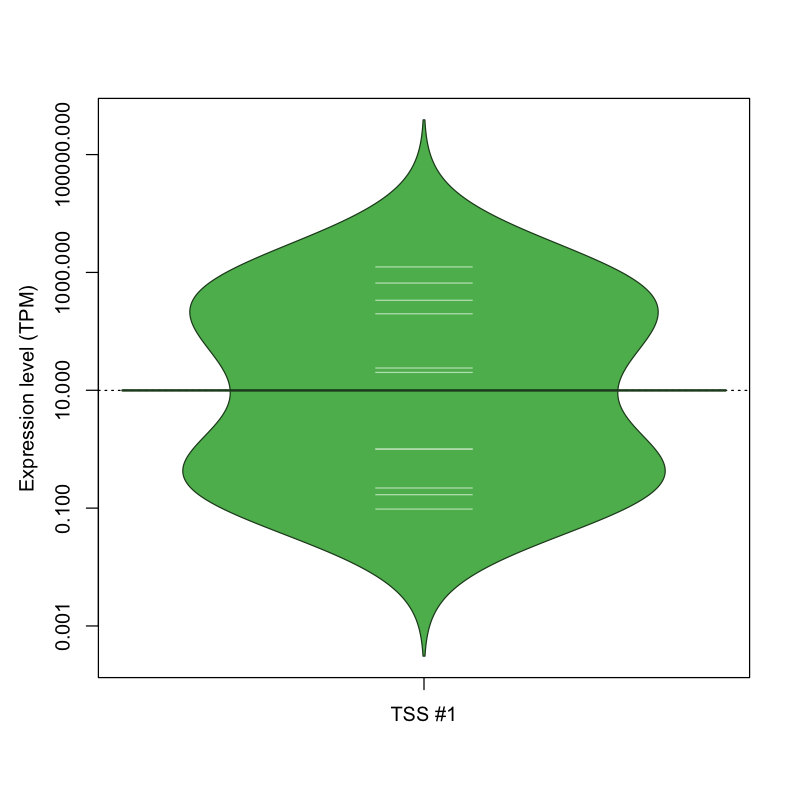
The graphical summary, for retro_mmus_119 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_119 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_119 has 5 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Homo sapiens | retro_hsap_70 |
| Callithrix jacchus | retro_cjac_125 |
| Rattus norvegicus | retro_rnor_34 |
| Sus scrofa | retro_sscr_59 |
| Canis familiaris | retro_cfam_19 |
Parental genes homology:
Parental genes homology involve 14 parental genes, and 25 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000007633 | 3 retrocopies | |
| Cavia porcellus | ENSCPOG00000003261 | 2 retrocopies | |
| Echinops telfairi | ENSETEG00000008279 | 2 retrocopies | |
| Felis catus | ENSFCAG00000004264 | 1 retrocopy | |
| Homo sapiens | ENSG00000198814 | 4 retrocopies | |
| Loxodonta africana | ENSLAFG00000007632 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000002417 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000025059 | 2 retrocopies |
retro_mmus_119 , retro_mmus_97,
|
| Nomascus leucogenys | ENSNLEG00000010102 | 2 retrocopies | |
| Oryctolagus cuniculus | ENSOCUG00000013023 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000034116 | 2 retrocopies | |
| Sarcophilus harrisii | ENSSHAG00000006470 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000008732 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000008249 | 1 retrocopy |