RetrogeneDB ID: | retro_hsap_4240 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 9:32566225..32566976(-) | ||
| Located in intron of: | ENSG00000165264 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000232303 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | DFFB | ||
| Ensembl ID: | ENSG00000169598 | ||
| Aliases: | DFFB, CAD, CPAN, DFF-40, DFF2, DFF40 | ||
| Description: | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) [Source:HGNC Symbol;Acc:2773] |

Retrocopy-Parental alignment summary:
>retro_hsap_4240
AGCGTCCTCGACGATGCCGAGCTGGTGCTGCTCACCTCACGCCAGGCCTGGCAGGGCTATGTGAGTGACATCGGGTGCTT
CCTCAGTGCTTTTCGCCAGCCGCACGCGTGGCTCATCCAGGCCGCCCGGCAGCTACTGTGTGATGAGCAGGCCCCCACAG
AGGCAGAGACTGCTGGCCGACCTCCTGTACAACGTCAGCCAGAACGTCGCAGCCGACACCCGGGCTGAGGACCCGCTGTG
GTTTGAAGGCTTGGAGTCCCGATTTTGGAATAAGTCTGGCTATCTGAGATACAGCTGTGAAAGCCGGATCCGGAGTTACC
TGAGAGAGGTGAGCTCCTGCTCCTCCATGGTGGGTGCAGAGGCTCAGGAGGAATTCCTGCGGGTCCTCCGCTCCATGTGC
CAGAAGCTCCAGTCTGTGCAGTACGACGGCAGCTATACAACAGAGGAGCCAAGGGTGGCAGCCGTCTCTGCACACCGGAA
GGCTAGTTCTCCTGCCAGGGTCCCTTCGACATGGACGGCTGCTTGTCAAGATACTCCATCAACCCCTACAGTAACAGGGA
GAGCAGGCTCCTCTTCTGGACCTGGAACCTGGATCACATAGTGGAAACAAAAACGCACCATCATTCCTACACTGGTGGAA
GCAGTTAAAGAACAAGATGGAAGAGAAGTGGACTGGGAATATTTTTATGGCCTGCTTTCTACCTCAGAGAACCTAAAATT
AGTGTTGTCTGCCATAAGAAAACCACCCACA
AGCGTCCTCGACGATGCCGAGCTGGTGCTGCTCACCTCACGCCAGGCCTGGCAGGGCTATGTGAGTGACATCGGGTGCTT
CCTCAGTGCTTTTCGCCAGCCGCACGCGTGGCTCATCCAGGCCGCCCGGCAGCTACTGTGTGATGAGCAGGCCCCCACAG
AGGCAGAGACTGCTGGCCGACCTCCTGTACAACGTCAGCCAGAACGTCGCAGCCGACACCCGGGCTGAGGACCCGCTGTG
GTTTGAAGGCTTGGAGTCCCGATTTTGGAATAAGTCTGGCTATCTGAGATACAGCTGTGAAAGCCGGATCCGGAGTTACC
TGAGAGAGGTGAGCTCCTGCTCCTCCATGGTGGGTGCAGAGGCTCAGGAGGAATTCCTGCGGGTCCTCCGCTCCATGTGC
CAGAAGCTCCAGTCTGTGCAGTACGACGGCAGCTATACAACAGAGGAGCCAAGGGTGGCAGCCGTCTCTGCACACCGGAA
GGCTAGTTCTCCTGCCAGGGTCCCTTCGACATGGACGGCTGCTTGTCAAGATACTCCATCAACCCCTACAGTAACAGGGA
GAGCAGGCTCCTCTTCTGGACCTGGAACCTGGATCACATAGTGGAAACAAAAACGCACCATCATTCCTACACTGGTGGAA
GCAGTTAAAGAACAAGATGGAAGAGAAGTGGACTGGGAATATTTTTATGGCCTGCTTTCTACCTCAGAGAACCTAAAATT
AGTGTTGTCTGCCATAAGAAAACCACCCACA
ORF - retro_hsap_4240 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 81.42 % |
| Parental protein coverage: | 73.96 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 3 |
Retrocopy - Parental Gene Alignment:
| Parental | SVPDNAELVLLTLGQAWQGYVSDIRRFLSAFHEPQVGLIQAAQQLLCDEQAP-QRQRLLADLLHNVSQNI |
| SV.D.AELVLLT..QAWQGYVSDI..FLSAF..P...LIQAA.QLLCDEQAP.QRQRLLADLL.NVSQN. | |
| Retrocopy | SVLDDAELVLLTSRQAWQGYVSDIGCFLSAFRQPHAWLIQAARQLLCDEQAP>QRQRLLADLLYNVSQNV |
| Parental | AAETRAEDPPWFEGLESRFQSKSGYLRYSCESRIRSYLREVSSYPSTVGAEAQEEFLRVLGSMCQRLRSM |
| AA.TRAEDP.WFEGLESRF..KSGYLRYSCESRIRSYLREVSS..S.VGAEAQEEFLRVL.SMCQ.L.S. | |
| Retrocopy | AADTRAEDPLWFEGLESRFWNKSGYLRYSCESRIRSYLREVSSCSSMVGAEAQEEFLRVLRSMCQKLQSV |
| Parental | QYNGSY-FDRGAKGGSRLCTPEGWFSCQGPFDMDSCLSRHSINPYSNRESRILFSTWNLDHIIE-KKRTI |
| QY.GSY...RGAKGGSRLCTPEG.FSCQGPFDMD.CLSR.SINPYSNRESR.LF.TWNLDHI.E..KRTI | |
| Retrocopy | QYDGSY<YNRGAKGGSRLCTPEG*FSCQGPFDMDGCLSRYSINPYSNRESRLLFWTWNLDHIVE>QKRTI |
| Parental | IPTLVEAIKEQDGREVDWEYFYGLLFTSENLKLVHIVCHKKTT |
| IPTLVEA.KEQDGREVDWEYFYGLL.TSENLKLV.....K..T | |
| Retrocopy | IPTLVEAVKEQDGREVDWEYFYGLLSTSENLKLVLSAIRKPPT |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .02 RPM | 2 .03 RPM |
| bodymap2_adrenal | 0 .70 RPM | 5 .49 RPM |
| bodymap2_brain | 0 .28 RPM | 3 .52 RPM |
| bodymap2_breast | 0 .33 RPM | 3 .37 RPM |
| bodymap2_colon | 0 .39 RPM | 1 .33 RPM |
| bodymap2_heart | 0 .20 RPM | 1 .57 RPM |
| bodymap2_kidney | 0 .50 RPM | 2 .13 RPM |
| bodymap2_liver | 0 .09 RPM | 1 .12 RPM |
| bodymap2_lung | 0 .00 RPM | 1 .35 RPM |
| bodymap2_lymph_node | 0 .64 RPM | 12 .65 RPM |
| bodymap2_ovary | 0 .54 RPM | 6 .09 RPM |
| bodymap2_prostate | 0 .17 RPM | 5 .96 RPM |
| bodymap2_skeletal_muscle | 0 .02 RPM | 0 .33 RPM |
| bodymap2_testis | 0 .26 RPM | 2 .04 RPM |
| bodymap2_thyroid | 0 .30 RPM | 2 .11 RPM |
| bodymap2_white_blood_cells | 0 .37 RPM | 7 .52 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_4240 was not detected
No EST(s) were mapped for retro_hsap_4240 retrocopy.
No TSS is located nearby retro_hsap_4240 retrocopy 5' end.
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | CAGAGGCGTTGAGTACATCT (20 nt long) |
| Reverse primer: | TCCTCTTCAACCTTGTCTGGG (21 nt long) |
| Anneling temperature: | 60 °C |
| (Expected) product size: | 976 |
| Electrophoresis gel image: | 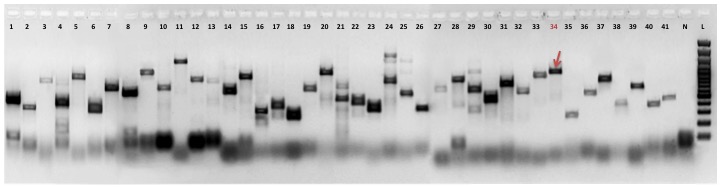 |
| Additional comment: | Red arrow indicates PCR product of retrogene in pooled cDNA from 16 human tissues (Clontech). Red number of lane indicates particular retrogene; L - GeneRuler 100 bp DNA Ladder (Thermo Fisher Scientific); N - No template control (water instead of cDNA) |
Retrocopy orthology:
Retrocopy retro_hsap_4240 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_2841 |
| Callithrix jacchus | retro_cjac_616 |
Parental genes homology:
Parental genes homology involve 5 parental genes, and 9 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000002834 | 1 retrocopy | |
| Homo sapiens | ENSG00000169598 | 1 retrocopy |
retro_hsap_4240 ,
|
| Gorilla gorilla | ENSGGOG00000013452 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000015174 | 1 retrocopy | |
| Ornithorhynchus anatinus | ENSOANG00000013652 | 5 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .53 RPM |
| CEU_NA11843 | 0 .45 RPM |
| CEU_NA11930 | 0 .49 RPM |
| CEU_NA12004 | 0 .42 RPM |
| CEU_NA12400 | 0 .25 RPM |
| CEU_NA12751 | 0 .78 RPM |
| CEU_NA12760 | 0 .67 RPM |
| CEU_NA12827 | 0 .47 RPM |
| CEU_NA12872 | 0 .49 RPM |
| CEU_NA12873 | 0 .22 RPM |
| FIN_HG00183 | 0 .36 RPM |
| FIN_HG00277 | 0 .63 RPM |
| FIN_HG00315 | 0 .72 RPM |
| FIN_HG00321 | 0 .57 RPM |
| FIN_HG00328 | 0 .66 RPM |
| FIN_HG00338 | 0 .47 RPM |
| FIN_HG00349 | 0 .20 RPM |
| FIN_HG00375 | 0 .81 RPM |
| FIN_HG00377 | 0 .53 RPM |
| FIN_HG00378 | 0 .70 RPM |
| GBR_HG00099 | 0 .49 RPM |
| GBR_HG00111 | 0 .62 RPM |
| GBR_HG00114 | 0 .39 RPM |
| GBR_HG00119 | 0 .50 RPM |
| GBR_HG00131 | 0 .52 RPM |
| GBR_HG00133 | 0 .72 RPM |
| GBR_HG00134 | 0 .87 RPM |
| GBR_HG00137 | 0 .41 RPM |
| GBR_HG00142 | 0 .42 RPM |
| GBR_HG00143 | 0 .26 RPM |
| TSI_NA20512 | 0 .45 RPM |
| TSI_NA20513 | 0 .87 RPM |
| TSI_NA20518 | 0 .47 RPM |
| TSI_NA20532 | 0 .54 RPM |
| TSI_NA20538 | 0 .32 RPM |
| TSI_NA20756 | 0 .35 RPM |
| TSI_NA20765 | 0 .70 RPM |
| TSI_NA20771 | 0 .34 RPM |
| TSI_NA20786 | 0 .37 RPM |
| TSI_NA20798 | 0 .78 RPM |
| YRI_NA18870 | 0 .85 RPM |
| YRI_NA18907 | 0 .42 RPM |
| YRI_NA18916 | 0 .76 RPM |
| YRI_NA19093 | 0 .94 RPM |
| YRI_NA19099 | 0 .40 RPM |
| YRI_NA19114 | 0 .81 RPM |
| YRI_NA19118 | 0 .50 RPM |
| YRI_NA19213 | 0 .60 RPM |
| YRI_NA19214 | 0 .93 RPM |
| YRI_NA19223 | 1 .13 RPM |