RetrogeneDB ID: | retro_mmus_205 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:186967875..186969858(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000039224 | |
| Aliases: | D1Pas1, Pl10 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ddx3x | ||
| Ensembl ID: | ENSMUSG00000000787 | ||
| Aliases: | Ddx3x, D1Pas1-rs2, Ddx3, Fin14 | ||
| Description: | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked [Source:MGI Symbol;Acc:MGI:103064] |

Retrocopy-Parental alignment summary:
>retro_mmus_205
ATGAGTCATGTGGCAGAGGAAGATGAGCTCGGGCTGGACCAGCAGTTGGCTGGCCTAGACCTGACGTCTCGGGACAGCCA
GAGCGGGGGCAGCACAGCCAGCAAAGGGCGCTACATCCCCCCTCACCTGCGGAACCGGGAGGCCGCGAAGGCGTTCTACG
ACAAAGACGGCTCACGGTGGAGTAAAGACAAGGATGCCTACAGCAGCTTCGGGTCCCGCAGCGACACGAGAGCCAAGTCC
AGTTTCTTCAGTGACCGCGGCGGAAGCGGCTCCAGGGGAAGGTTTGACGAGCGCGGGCGGAGTGACTACGAGAGTGTTGG
CAGCCGGGGAGGTAGAAGTGGCTTTGGCAAATTTGAGCGGGGCGGGAACAGTCGCTGGTGTGACAAAGCAGACGAGGATG
ACTGGTCGAAACCGCTCCCACCAAGCGAACGCTTGGAACAGGAACTCTTTTCCGGAGGCAACACTGGGATTAACTTTGAG
AAATATGATGACATCCCGGTTGAGGCGACAGGGAATAACTGCCCTCCACATATTGAAAGTTTCAGCGATGTCGAGATGGG
AGAGATTATTATGGGAAACATCGAGCTCACTCGTTACACACGCCCAACTCCTGTGCAAAAGCATGCTATTCCTATTATCA
AGGAGAAAAGAGACTTGATGGCTTGTGCCCAAACGGGGTCTGGAAAAACAGCAGCTTTTCTTCTACCTATCTTGAGTCAG
ATCTATACGGATGGTCCAGGTGAGGCTTTGAGGGCCATGAAGGAGAATGGAAAGTATGGGCGTCGCAAACAGTATCCAAT
CTCCTTGGTCTTGGCTCCGACCAGAGAATTGGCCGTGCAGATCTATGAGGAGGCCAGGAAATTCTCATACCGCTCTCGAG
TTCGTCCTTGTGTGGTTTATGGTGGAGCTGATATTGGGCAGCAGATTCGAGACTTGGAACGTGGCTGTCACTTGTTAGTA
GCTACTCCAGGACGGCTAGTAGATATGATGGAGAGAGGAAAGATTGGACTGGACTTCTGCAAATACTTGGTGTTAGATGA
AGCAGATCGAATGTTGGACATGGGCTTTGAGCCTCAGATACGAAGAATAGTTGAGCAAGACACCATGCCTCCAAAAGGTG
TCCGCCACACTATGATGTTTAGTGCTACCTTTCCTAAGGAAATACAGATGCTGGCTCGGGATTTCTTGGATGAATATATC
TTTCTGGCCGTAGGAAGAGTTGGGTCTACTTCTGAGAACATCACACAGAAAGTCGTGTGGGTGGAAGAAGCAGACAAGCG
GTCATTTCTGCTTGACCTCCTGAATGCAACGGGCAAGGATTCCCTGATATTGGTGTTTGTGGAAACCAAAAAGGGTGCAG
ATTCTCTGGAGGACTTCTTATACCACGAAGGATATGCTTGTACCAGTATCCATGGAGACCGATCTCAGCGAGATAGGGAA
GAGGCCCTTCACCAATTCCGATCAGGAAAAAGCCCAATTCTCGTGGCTACAGCAGTAGCTGCAAGAGGACTGGATATTTC
CAATGTGAAACATGTCATCAATTTCGACTTGCCTAGTGACATCGAAGAATATGTGCATCGCATTGGCCGTACAGGTCGTG
TTGGAAACCTTGGCCTTGCCACCTCATTCTTCAACGAGAGGAACATAAATATTACCAAAGATTTATTGGATCTTCTTGTT
GAAGCAAAACAAGAAGTGCCATCTTGGCTAGAAAACATGGCTTTTGAACACCATTACAAGGGTGGCAGTCGTGGACGGTC
CAAGAGCAGATTTAGTGGAGGATTTGGTGCTAGAGACTATCGACAAAGTAGCGGTGCCAGCAGTTCCAGCTTCAGCAGTG
GCCGTGCTAGCAACAGTCGCAGCGGTGGAGGCAGCCATGGCAGCAGCAGAGGGTTTGGTGGAGGTAGCTATGGAGGCTTT
TACAACAGTGATGGATATGGAGGAAATTATAGCTCCCAGGGGGTTGACTGGTGGGGTAACTGA
ATGAGTCATGTGGCAGAGGAAGATGAGCTCGGGCTGGACCAGCAGTTGGCTGGCCTAGACCTGACGTCTCGGGACAGCCA
GAGCGGGGGCAGCACAGCCAGCAAAGGGCGCTACATCCCCCCTCACCTGCGGAACCGGGAGGCCGCGAAGGCGTTCTACG
ACAAAGACGGCTCACGGTGGAGTAAAGACAAGGATGCCTACAGCAGCTTCGGGTCCCGCAGCGACACGAGAGCCAAGTCC
AGTTTCTTCAGTGACCGCGGCGGAAGCGGCTCCAGGGGAAGGTTTGACGAGCGCGGGCGGAGTGACTACGAGAGTGTTGG
CAGCCGGGGAGGTAGAAGTGGCTTTGGCAAATTTGAGCGGGGCGGGAACAGTCGCTGGTGTGACAAAGCAGACGAGGATG
ACTGGTCGAAACCGCTCCCACCAAGCGAACGCTTGGAACAGGAACTCTTTTCCGGAGGCAACACTGGGATTAACTTTGAG
AAATATGATGACATCCCGGTTGAGGCGACAGGGAATAACTGCCCTCCACATATTGAAAGTTTCAGCGATGTCGAGATGGG
AGAGATTATTATGGGAAACATCGAGCTCACTCGTTACACACGCCCAACTCCTGTGCAAAAGCATGCTATTCCTATTATCA
AGGAGAAAAGAGACTTGATGGCTTGTGCCCAAACGGGGTCTGGAAAAACAGCAGCTTTTCTTCTACCTATCTTGAGTCAG
ATCTATACGGATGGTCCAGGTGAGGCTTTGAGGGCCATGAAGGAGAATGGAAAGTATGGGCGTCGCAAACAGTATCCAAT
CTCCTTGGTCTTGGCTCCGACCAGAGAATTGGCCGTGCAGATCTATGAGGAGGCCAGGAAATTCTCATACCGCTCTCGAG
TTCGTCCTTGTGTGGTTTATGGTGGAGCTGATATTGGGCAGCAGATTCGAGACTTGGAACGTGGCTGTCACTTGTTAGTA
GCTACTCCAGGACGGCTAGTAGATATGATGGAGAGAGGAAAGATTGGACTGGACTTCTGCAAATACTTGGTGTTAGATGA
AGCAGATCGAATGTTGGACATGGGCTTTGAGCCTCAGATACGAAGAATAGTTGAGCAAGACACCATGCCTCCAAAAGGTG
TCCGCCACACTATGATGTTTAGTGCTACCTTTCCTAAGGAAATACAGATGCTGGCTCGGGATTTCTTGGATGAATATATC
TTTCTGGCCGTAGGAAGAGTTGGGTCTACTTCTGAGAACATCACACAGAAAGTCGTGTGGGTGGAAGAAGCAGACAAGCG
GTCATTTCTGCTTGACCTCCTGAATGCAACGGGCAAGGATTCCCTGATATTGGTGTTTGTGGAAACCAAAAAGGGTGCAG
ATTCTCTGGAGGACTTCTTATACCACGAAGGATATGCTTGTACCAGTATCCATGGAGACCGATCTCAGCGAGATAGGGAA
GAGGCCCTTCACCAATTCCGATCAGGAAAAAGCCCAATTCTCGTGGCTACAGCAGTAGCTGCAAGAGGACTGGATATTTC
CAATGTGAAACATGTCATCAATTTCGACTTGCCTAGTGACATCGAAGAATATGTGCATCGCATTGGCCGTACAGGTCGTG
TTGGAAACCTTGGCCTTGCCACCTCATTCTTCAACGAGAGGAACATAAATATTACCAAAGATTTATTGGATCTTCTTGTT
GAAGCAAAACAAGAAGTGCCATCTTGGCTAGAAAACATGGCTTTTGAACACCATTACAAGGGTGGCAGTCGTGGACGGTC
CAAGAGCAGATTTAGTGGAGGATTTGGTGCTAGAGACTATCGACAAAGTAGCGGTGCCAGCAGTTCCAGCTTCAGCAGTG
GCCGTGCTAGCAACAGTCGCAGCGGTGGAGGCAGCCATGGCAGCAGCAGAGGGTTTGGTGGAGGTAGCTATGGAGGCTTT
TACAACAGTGATGGATATGGAGGAAATTATAGCTCCCAGGGGGTTGACTGGTGGGGTAACTGA
ORF - retro_mmus_205 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 94.12 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSHVAVENALGLDQQFAGLDLNSSDNQSGGSTASKGRYIPPHLRNREATKGFYDKDSSGWSSSKDKDAYS |
| MSHVA.E..LGLDQQ.AGLDL.S.D.QSGGSTASKGRYIPPHLRNREA.K.FYDKD.S.WS..KDKDAYS | |
| Retrocopy | MSHVAEEDELGLDQQLAGLDLTSRDSQSGGSTASKGRYIPPHLRNREAAKAFYDKDGSRWS--KDKDAYS |
| Parental | SFGSRGDSRGKSSFFGDRG-SGSRGRFDDRGRGDYDGIGGRGDRSGFGKFERGGNSRWCDKSDEDDWSKP |
| SFGSR.D.R.KSSFF.DRG.SGSRGRFD.RGR.DY...G.RG.RSGFGKFERGGNSRWCDK.DEDDWSKP | |
| Retrocopy | SFGSRSDTRAKSSFFSDRGGSGSRGRFDERGRSDYESVGSRGGRSGFGKFERGGNSRWCDKADEDDWSKP |
| Parental | LPPSERLEQELFSGGNTGINFEKYDDIPVEATGNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKH |
| LPPSERLEQELFSGGNTGINFEKYDDIPVEATGNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKH | |
| Retrocopy | LPPSERLEQELFSGGNTGINFEKYDDIPVEATGNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKH |
| Parental | AIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYADGPGEALRAMKENGRYGRRKQYPISLVLAPTRELA |
| AIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIY.DGPGEALRAMKENG.YGRRKQYPISLVLAPTRELA | |
| Retrocopy | AIPIIKEKRDLMACAQTGSGKTAAFLLPILSQIYTDGPGEALRAMKENGKYGRRKQYPISLVLAPTRELA |
| Parental | VQIYEEARKFSYRSRVRPCVVYGGAEIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLDFCKYLVLDEA |
| VQIYEEARKFSYRSRVRPCVVYGGA.IGQQIRDLERGCHLLVATPGRLVDMMERGKIGLDFCKYLVLDEA | |
| Retrocopy | VQIYEEARKFSYRSRVRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLDFCKYLVLDEA |
| Parental | DRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDEYIFLAVGRVGSTSENITQKV |
| DRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDEYIFLAVGRVGSTSENITQKV | |
| Retrocopy | DRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDEYIFLAVGRVGSTSENITQKV |
| Parental | VWVEEIDKRSFLLDLLNATGKDSLTLVFVETKKGADSLEDFLYHEGYACTSIHGDRSQRDREEALHQFRS |
| VWVEE.DKRSFLLDLLNATGKDSL.LVFVETKKGADSLEDFLYHEGYACTSIHGDRSQRDREEALHQFRS | |
| Retrocopy | VWVEEADKRSFLLDLLNATGKDSLILVFVETKKGADSLEDFLYHEGYACTSIHGDRSQRDREEALHQFRS |
| Parental | GKSPILVATAVAARGLDISNVKHVINFDLPSDIEEYVHRIGRTGRVGNLGLATSFFNERNINITKDLLDL |
| GKSPILVATAVAARGLDISNVKHVINFDLPSDIEEYVHRIGRTGRVGNLGLATSFFNERNINITKDLLDL | |
| Retrocopy | GKSPILVATAVAARGLDISNVKHVINFDLPSDIEEYVHRIGRTGRVGNLGLATSFFNERNINITKDLLDL |
| Parental | LVEAKQEVPSWLENMAFEHHYKGSSRGRSKSSRFSGGFGARDYRQSSGASSSSFSSSRASSSRSGGGGHG |
| LVEAKQEVPSWLENMAFEHHYKG.SRGRSK.SRFSGGFGARDYRQSSGASSSSFSS.RAS.SRSGGG.HG | |
| Retrocopy | LVEAKQEVPSWLENMAFEHHYKGGSRGRSK-SRFSGGFGARDYRQSSGASSSSFSSGRASNSRSGGGSHG |
| Parental | GSRGFGGGGYGGFYNSDGYGGNYNSQGVDWWGN |
| .SRGFGGG.YGGFYNSDGYGGNY.SQGVDWWGN | |
| Retrocopy | SSRGFGGGSYGGFYNSDGYGGNYSSQGVDWWGN |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 210 .69 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 222 .97 RPM |
| SRP007412_heart | 0 .06 RPM | 218 .81 RPM |
| SRP007412_kidney | 0 .00 RPM | 285 .76 RPM |
| SRP007412_liver | 0 .08 RPM | 248 .10 RPM |
| SRP007412_testis | 254 .77 RPM | 34 .12 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_205 was not detected
No EST(s) were mapped for retro_mmus_205 retrocopy.
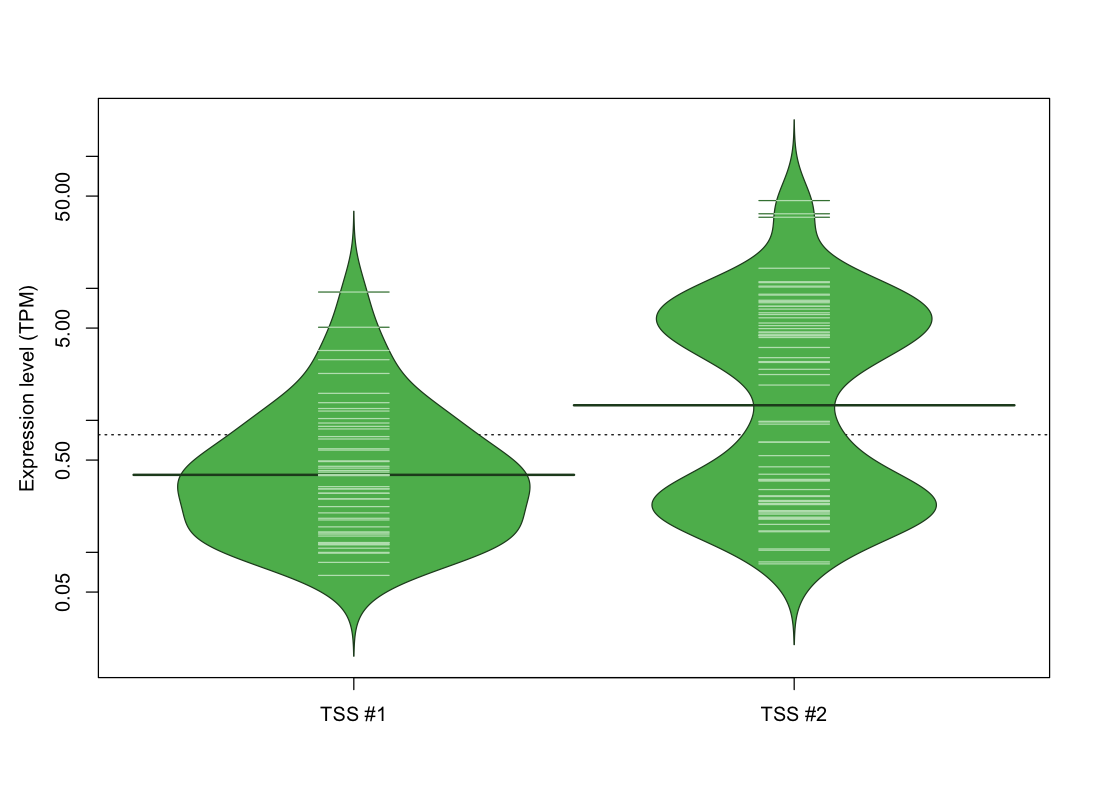
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_73153 | 1019 libraries | 43 libraries | 8 libraries | 2 libraries | 0 libraries |
| TSS #2 | TSS_73154 | 1000 libraries | 36 libraries | 15 libraries | 13 libraries | 8 libraries |
The graphical summary, for retro_mmus_205 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_205 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_205 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 14 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000035907 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000009050 | 2 retrocopies | |
| Cavia porcellus | ENSCPOG00000013557 | 2 retrocopies | |
| Equus caballus | ENSECAG00000009901 | 1 retrocopy | |
| Homo sapiens | ENSG00000215301 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000017569 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000000787 | 1 retrocopy |
retro_mmus_205 ,
|
| Ornithorhynchus anatinus | ENSOANG00000011274 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000020257 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000022488 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000023383 | 1 retrocopy |