RetrogeneDB ID: | retro_mmus_22 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 19:24476501..24477356(-) | ||
| Located in intron of: | ENSMUSG00000024867 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000074922 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Fam122b | ||
| Ensembl ID: | ENSMUSG00000036022 | ||
| Aliases: | Fam122b, 4632404H22Rik, AI256344, SPACIA2 | ||
| Description: | family with sequence similarity 122, member B [Source:MGI Symbol;Acc:MGI:1926005] |

Retrocopy-Parental alignment summary:
>retro_mmus_22
ATGGCTCAGGAGAAGATGGAGCTGGACCTGGAGTTGCCCGCGGGCGCGAGCCCGGCGGAGGGCGGCGGCCCTGGCGGCGG
GGGCCTGCGGAGGTCTAACAGCGCCCCCCTGATCCACGGCCTCAGCGACTCGTCGCCGGTGTTCCAGGCCGAGGCGCCCA
GCGCCAGACGGAACAGCACGACGTTCCCGAGCCGCCACGGCCTGCTGCTCCCGGCCTCGCCGGTCCGAATGCACAGCAGC
CGCCTGCACCAGATCAAACAGGAGGAGGGCATGGACCTGATCAACCGAGAGACCGTTCACGAGCGCGAGGTGCAGACCGC
AATGCAGATAAGCCACTCCTGGGAGGAAAGTTTCAGCCTGAGTGACAACGACGTGGAGAAATCCGCCTCCCCGAAGCGCA
TCGATTTCATTCCGGTGTCTCCGGCACCGTCCCCGACCCGGGGAATTGGGAAGCAGTGTTTCTCACCGTCCTTGCAAAGT
TTTGTGAGTAGCAACGGACTGCCTCCAAGTCCCATTCCCAGCCCAACCACCCGATTTACCACCCGGAGAAGCCAGAGTCC
CATCAACTGCATTAGACCAAGTGTTCTTGGACCATTGAAAAGAAAATGTGAAATGGAAACTGATTATCAGCCAAAGAGAT
TTTTCCAGGGCATCACCAACATGCTGTCTTCTGACGTTGCACAGCTGTCAGACCCCGGTGTGTGCGTATCTTCCGATACC
CTGGATGGAAACAGCAGCAGTGCCGGATCTTCCTGTAACTCACCAGCGAAAGTCAGCACTACCACCGACTCTCCTGTGTC
GCCCGCCCAAGCAGCCTCCCCCTTTATTCCAGTAGATGAACTTTCCTCAAAGTGA
ORF - retro_mmus_22 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 63.5 % |
| Parental protein coverage: | 76.95 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | EEGLDMMNRETTNEREAQAGMQISQSWDESLSLSDSDFDKPEKLYSPKRIDFTPVSPAPSPTRGFGKQCL |
| EEG.D..NRET..ERE.Q..MQIS.SW.ES.SLSD.D....EK..SPKRIDF.PVSPAPSPTRG.GKQC. | |
| Retrocopy | EEGMDLINRETVHEREVQTAMQISHSWEESFSLSDNDV---EKSASPKRIDFIPVSPAPSPTRGIGKQCF |
| Parental | SPSLQMFVSSSGMPPSPVLNP-RHFSSRRSQSPVKCIRPSVLGPLKRKGEMEMESQPKRPFQGTTSMLST |
| SPSLQ.FVSS.G.PPSP...P...F..RRSQSP..CIRPSVLGPLKRK.EME...QPKR.FQG.T.MLS. | |
| Retrocopy | SPSLQSFVSSNGLPPSPIPSPTTRFTTRRSQSPINCIRPSVLGPLKRKCEMETDYQPKRFFQGITNMLSS |
| Parental | NPAQLSDFSSC--SDILDGSSISSGLSSDSLATGSAPAESPVACSNSCSPFILMDDLSPK |
| ..AQLSD...C..SD.LDG.S.S.G.S..S.A..S....SPV......SPFI..D.LS.K | |
| Retrocopy | DVAQLSDPGVCVSSDTLDGNSSSAGSSCNSPAKVSTTTDSPVSPAQAASPFIPVDELSSK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 6 .59 RPM | 5 .37 RPM |
| SRP007412_cerebellum | 7 .34 RPM | 4 .56 RPM |
| SRP007412_heart | 11 .89 RPM | 4 .51 RPM |
| SRP007412_kidney | 9 .70 RPM | 7 .47 RPM |
| SRP007412_liver | 6 .31 RPM | 3 .00 RPM |
| SRP007412_testis | 8 .23 RPM | 2 .88 RPM |
RNA Polymerase II activity may be related with retro_mmus_22 in 3 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 19:24476947..24477763 |
| ENCFF001YIJ | POLR2A | 19:24476454..24479066 |
| ENCFF001YKA | POLR2A | 19:24474297..24479094 |
No EST(s) were mapped for retro_mmus_22 retrocopy.
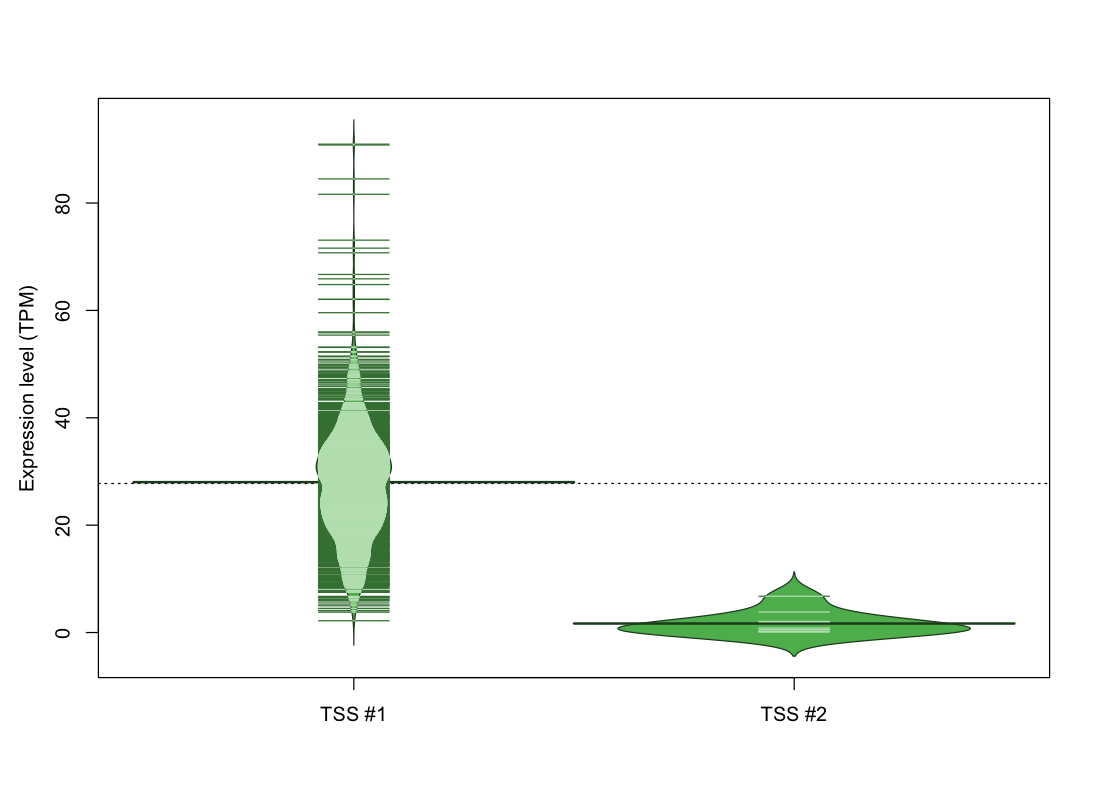
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_63200 | 6 libraries | 0 libraries | 6 libraries | 40 libraries | 1020 libraries |
| TSS #2 | TSS_63202 | 1062 libraries | 6 libraries | 3 libraries | 1 library | 0 libraries |
The graphical summary, for retro_mmus_22 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_22 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_22 has 6 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 14 parental genes, and 14 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000001311 | 1 retrocopy | |
| Bos taurus | ENSBTAG00000001886 | 1 retrocopy | |
| Canis familiaris | ENSCAFG00000018882 | 1 retrocopy | |
| Callithrix jacchus | ENSCJAG00000004313 | 1 retrocopy | |
| Cavia porcellus | ENSCPOG00000011218 | 1 retrocopy | |
| Equus caballus | ENSECAG00000018643 | 1 retrocopy | |
| Felis catus | ENSFCAG00000024842 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000007988 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000016948 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000012690 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000036022 | 1 retrocopy |
retro_mmus_22 ,
|
| Rattus norvegicus | ENSRNOG00000050146 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000012685 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000006261 | 1 retrocopy |