RetrogeneDB ID: | retro_mmus_2519 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 4:139345302..139346180(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000080708 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ccnd3 | ||
| Ensembl ID: | ENSMUSG00000034165 | ||
| Aliases: | Ccnd3, 9230106B05Rik, AA682053, AL024085, AW146355, C78795 | ||
| Description: | cyclin D3 [Source:MGI Symbol;Acc:MGI:88315] |

Retrocopy-Parental alignment summary:
>retro_mmus_2519
ATGGAGCTGCTATGTTGCGAGGGCACCCGGCACGCGCCCCGGGCCAGGCCCGACCCGCGGCTGCTTATGGACCAGTGTGT
CCTGCAGAGTTTACTCCGCCTGGAGGAGCGCTACGTGCCGCGATCCTCCTCCTTCCAGTGCATGCAAAAGAAGATCAAGC
CGCACATGCGGAAGATGCTGGCATACTGGATGCTGGAGGTGTGTGAGGAGCAGCGCTGCGAGGAGGATGTCTTCCATCTG
GCTATGATCTACCTGTATCGCTACCTGTCCTGCGTCCCCACCCGAAAGGCGCAATTGCAGCTTCTAGGTACCCGTCTGCC
TGTTGCTGGCCTCCAAGCTGCGCGAAACCACGCCCCTGACTATTGAGAAGCTTTGCATCTATTACGGACCAGGCTGTGGC
TCCATGGCAGTTGCGGGAGTGGGAGGTGCTGGTCCTGGGGAAGCTCAAGTGGGACCTGGCTGCTGTGATTGCGCACGACT
TCCTGGCCCTGATTCTGCACCGCCTGTCTCTGCCCAGTGGTTGGCAGGCCTTGGTCAAAAAGCATGCCCAGACTTTTTTG
GCCCTCTGTGCTACAGATTACACCTTTGCGATGTATCCTCCATCCATGATTGCCACAGGCAGCATTGGGGCGGCAGTGCT
AGGCCTAGGCGCCTGCTCTATGTCTGCGGATGAGCTCACAGAGTTGCTGGCCGGGATCACAGGCACTGAAGTGGACTGCC
TGCGTGCCTGTCAGGAGCAGATCGAAGCTGCCCTCAGGGAGAACCTCAGGGAAGCTGCTCAGACAGCCCCCAGCCCGGTG
TCCAAAGTCCCCCGGAGCTCTAGCAGCCAGGGGCCCGGTCAGACCAGCACTCCCACAGATGTCACAGCCATTCACCTG
ATGGAGCTGCTATGTTGCGAGGGCACCCGGCACGCGCCCCGGGCCAGGCCCGACCCGCGGCTGCTTATGGACCAGTGTGT
CCTGCAGAGTTTACTCCGCCTGGAGGAGCGCTACGTGCCGCGATCCTCCTCCTTCCAGTGCATGCAAAAGAAGATCAAGC
CGCACATGCGGAAGATGCTGGCATACTGGATGCTGGAGGTGTGTGAGGAGCAGCGCTGCGAGGAGGATGTCTTCCATCTG
GCTATGATCTACCTGTATCGCTACCTGTCCTGCGTCCCCACCCGAAAGGCGCAATTGCAGCTTCTAGGTACCCGTCTGCC
TGTTGCTGGCCTCCAAGCTGCGCGAAACCACGCCCCTGACTATTGAGAAGCTTTGCATCTATTACGGACCAGGCTGTGGC
TCCATGGCAGTTGCGGGAGTGGGAGGTGCTGGTCCTGGGGAAGCTCAAGTGGGACCTGGCTGCTGTGATTGCGCACGACT
TCCTGGCCCTGATTCTGCACCGCCTGTCTCTGCCCAGTGGTTGGCAGGCCTTGGTCAAAAAGCATGCCCAGACTTTTTTG
GCCCTCTGTGCTACAGATTACACCTTTGCGATGTATCCTCCATCCATGATTGCCACAGGCAGCATTGGGGCGGCAGTGCT
AGGCCTAGGCGCCTGCTCTATGTCTGCGGATGAGCTCACAGAGTTGCTGGCCGGGATCACAGGCACTGAAGTGGACTGCC
TGCGTGCCTGTCAGGAGCAGATCGAAGCTGCCCTCAGGGAGAACCTCAGGGAAGCTGCTCAGACAGCCCCCAGCCCGGTG
TCCAAAGTCCCCCGGAGCTCTAGCAGCCAGGGGCCCGGTCAGACCAGCACTCCCACAGATGTCACAGCCATTCACCTG
ORF - retro_mmus_2519 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.54 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MELLCCEGTRHAPRAGPDPRLLGDQRVLQSLLRLEERYVPRASYFQCVQKEIKPHMRKMLAYWMLEVCEE |
| MELLCCEGTRHAPRA.PDPRLL.DQ.VLQSLLRLEERYVPR.S.FQC.QK.IKPHMRKMLAYWMLEVCEE | |
| Retrocopy | MELLCCEGTRHAPRARPDPRLLMDQCVLQSLLRLEERYVPRSSSFQCMQKKIKPHMRKMLAYWMLEVCEE |
| Parental | QRCEEDVFPLAMNYLDRYLSCVPTRKAQLQLLGT-VCLLLASKLRETTPLTIEKLCIY-TDQAVAPWQLR |
| QRCEEDVF.LAM.YL.RYLSCVPTRKAQLQLLGT.VCLLLASKLRETTPLTIEKLCIY.TDQAVAPWQLR | |
| Retrocopy | QRCEEDVFHLAMIYLYRYLSCVPTRKAQLQLLGT>VCLLLASKLRETTPLTIEKLCIY>TDQAVAPWQLR |
| Parental | EWEVLVLGKLKWDLAAVIAHDFLALILHRLSLPSDRQALVKKHAQTFLALCATDYTFAMYPPSMIATGSI |
| EWEVLVLGKLKWDLAAVIAHDFLALILHRLSLPS..QALVKKHAQTFLALCATDYTFAMYPPSMIATGSI | |
| Retrocopy | EWEVLVLGKLKWDLAAVIAHDFLALILHRLSLPSGWQALVKKHAQTFLALCATDYTFAMYPPSMIATGSI |
| Parental | GAAVLGLGACSMSADELTELLAGITGTEVDCLRACQEQIEAALRESLREAAQTAPSPVPKAPRGSSSQGP |
| GAAVLGLGACSMSADELTELLAGITGTEVDCLRACQEQIEAALRE.LREAAQTAPSPV.K.PR.SSSQGP | |
| Retrocopy | GAAVLGLGACSMSADELTELLAGITGTEVDCLRACQEQIEAALRENLREAAQTAPSPVSKVPRSSSSQGP |
| Parental | SQTSTPTDVTAIHL |
| .QTSTPTDVTAIHL | |
| Retrocopy | GQTSTPTDVTAIHL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 12 .13 RPM |
| SRP007412_cerebellum | 0 .09 RPM | 18 .58 RPM |
| SRP007412_heart | 0 .00 RPM | 34 .65 RPM |
| SRP007412_kidney | 0 .08 RPM | 53 .13 RPM |
| SRP007412_liver | 0 .00 RPM | 17 .42 RPM |
| SRP007412_testis | 0 .05 RPM | 71 .76 RPM |
RNA Polymerase II actvity may be related with retro_mmus_2519 in 2 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001YIJ | POLR2A | 4:139345568..139346989 |
| ENCFF001YKA | POLR2A | 4:139345839..139347301 |
No EST(s) were mapped for retro_mmus_2519 retrocopy.
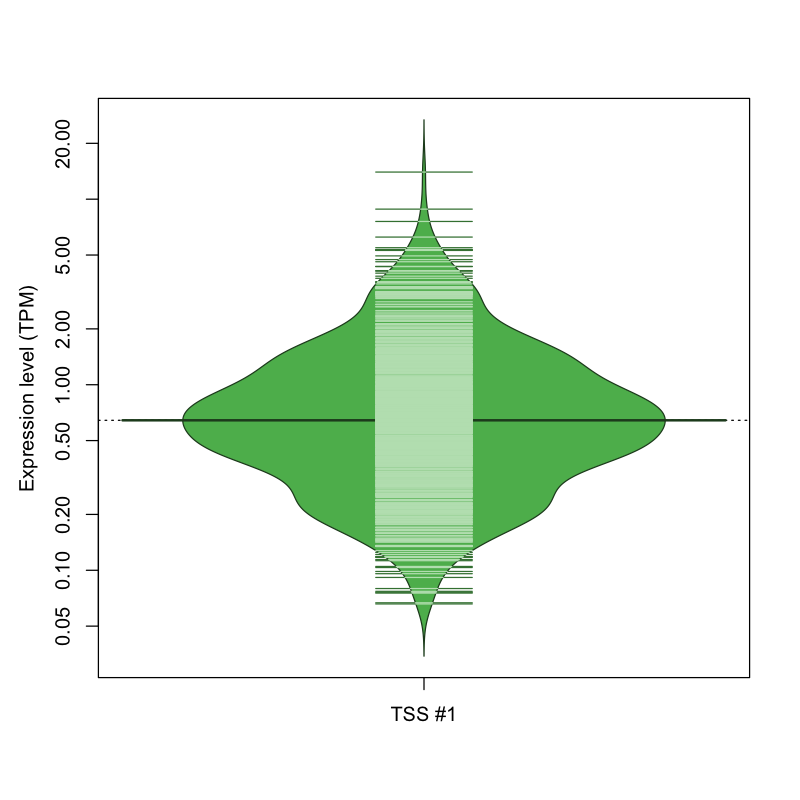
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_102265 | 347 libraries | 516 libraries | 200 libraries | 8 libraries | 1 library |
The graphical summary, for retro_mmus_2519 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2519 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2519 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 2 parental genes, and 6 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Mus musculus | ENSMUSG00000034165 | 1 retrocopy |
retro_mmus_2519 ,
|
| Mus musculus | ENSMUSG00000041431 | 5 retrocopies |