RetrogeneDB ID: | retro_mmus_2730 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 6:47045155..47046286(+) | ||
| Located in intron of: | ENSMUSG00000039419 | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ccni | ||
| Ensembl ID: | ENSMUSG00000063015 | ||
| Aliases: | None | ||
| Description: | cyclin I [Source:MGI Symbol;Acc:MGI:1341077] |

Retrocopy-Parental alignment summary:
>retro_mmus_2730
ATGAAGATTCCAGGACCTTTGGAAAGCCAGAGATTGTTTTTTTGTTGGAAAGGGCAATCTCCAGGGAAGCCCAGATGTGG
AAGGTGAATGTGCCGAAAATACCTACAAATCAGAATGTTTCTCCATCCCAGAGAGATGAAGTAATTCAATGGTTGGCCAA
ATTCAAATACCAGTTCAACCTCTATCCAGAAACATTTGCTCTGGCAAGCAGTCTTTTGGATAGGATTTTAGCTACAGTAA
AAGCCCATCCAAAATATTTGAATTATATTGCAATCAGCTGGTTTTTTCTGGCTGCTAAGACTGTTGAGGAAGATGAGAAA
ATTCCAGTGCTAAAGGTATTGGCAAGAGACAGTTTCTGTGGGTGTTCCTCATCTGAGATTTTGAGAATGGAGAGAATTAT
TCTGGATAAATTGAATTGGGATCTTCACATGGCCACACCATTGGATTTTCTTCACATTTTCCATGCCATTGTGGTGTCAA
CTAGGCCTCAGTTACTTTTCAGTTGGCCCAAATTGAGCCCATCTCAACATTTGGTAATCCTGACCAAAAAGCTGCTTCCC
TGTATGGCCTGCAACCAACTTCTGCAGTTCAAAGGGTCCATGCTGGCTCTGGCCATGGTTAGTCTGGAAATGGAGAAACC
CATTCCTGATTGGCTTCCTCTTACGATTGAACTGCTTCAGGAAGCAGAGATGGGCAGCTCCCAGTAGATCCACTGCCGGG
GAGCTGGTGGCATATCACTGTTCTGCTCCGCAGTCTGCCCTGCCTCTAAATTCCGTTTATGTCTACCGTCCCCTCAAGCA
CACCCTGGTAACCTGTGACAAAGGAGCATTCACATTACATCCCTCCTCTGTCTCAGGCCCAGATTTCTCCAAGGACAAGA
GCAAGCCAGAAGTGCCAGTCCGAGGTCCAGCAGCCTTCCACCTGCATCTCCCCGCTGCCAGTGGGTGCAAGCAAACCTCT
GCTAAATGGAAAGTGGAGGAGATGGAGGTGGATGACTTCTCCAATGGGATCAAGCCGCTCTATAATGAGGACAATGGTCC
TGAGAATGTGGGTTCTGTATGTGGCACTGATTTATCAAGGCAAGAGGGCCATGCTTCCCCCTGTCCACCTTTGCAGCCTG
TTTCTGTCATG
ATGAAGATTCCAGGACCTTTGGAAAGCCAGAGATTGTTTTTTTGTTGGAAAGGGCAATCTCCAGGGAAGCCCAGATGTGG
AAGGTGAATGTGCCGAAAATACCTACAAATCAGAATGTTTCTCCATCCCAGAGAGATGAAGTAATTCAATGGTTGGCCAA
ATTCAAATACCAGTTCAACCTCTATCCAGAAACATTTGCTCTGGCAAGCAGTCTTTTGGATAGGATTTTAGCTACAGTAA
AAGCCCATCCAAAATATTTGAATTATATTGCAATCAGCTGGTTTTTTCTGGCTGCTAAGACTGTTGAGGAAGATGAGAAA
ATTCCAGTGCTAAAGGTATTGGCAAGAGACAGTTTCTGTGGGTGTTCCTCATCTGAGATTTTGAGAATGGAGAGAATTAT
TCTGGATAAATTGAATTGGGATCTTCACATGGCCACACCATTGGATTTTCTTCACATTTTCCATGCCATTGTGGTGTCAA
CTAGGCCTCAGTTACTTTTCAGTTGGCCCAAATTGAGCCCATCTCAACATTTGGTAATCCTGACCAAAAAGCTGCTTCCC
TGTATGGCCTGCAACCAACTTCTGCAGTTCAAAGGGTCCATGCTGGCTCTGGCCATGGTTAGTCTGGAAATGGAGAAACC
CATTCCTGATTGGCTTCCTCTTACGATTGAACTGCTTCAGGAAGCAGAGATGGGCAGCTCCCAGTAGATCCACTGCCGGG
GAGCTGGTGGCATATCACTGTTCTGCTCCGCAGTCTGCCCTGCCTCTAAATTCCGTTTATGTCTACCGTCCCCTCAAGCA
CACCCTGGTAACCTGTGACAAAGGAGCATTCACATTACATCCCTCCTCTGTCTCAGGCCCAGATTTCTCCAAGGACAAGA
GCAAGCCAGAAGTGCCAGTCCGAGGTCCAGCAGCCTTCCACCTGCATCTCCCCGCTGCCAGTGGGTGCAAGCAAACCTCT
GCTAAATGGAAAGTGGAGGAGATGGAGGTGGATGACTTCTCCAATGGGATCAAGCCGCTCTATAATGAGGACAATGGTCC
TGAGAATGTGGGTTCTGTATGTGGCACTGATTTATCAAGGCAAGAGGGCCATGCTTCCCCCTGTCCACCTTTGCAGCCTG
TTTCTGTCATG
ORF - retro_mmus_2730 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 91.82 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MKFPGPLENQRLSS-LLERAISREAQMWKVNVPKIPTNQNVSPSQRDEVIQWLAKLKYQFNLYPETFALA |
| MK.PGPLE.QRL...LLERAISREAQMWKVNVPKIPTNQNVSPSQRDEVIQWLAK.KYQFNLYPETFALA | |
| Retrocopy | MKIPGPLESQRLFF<LLERAISREAQMWKVNVPKIPTNQNVSPSQRDEVIQWLAKFKYQFNLYPETFALA |
| Parental | SSLLDRFLATVKAHPKYLNCIAISCFFLAAKTVEEDEKIPVLKVLARDSFCGCSSSEILRMERIILDKLN |
| SSLLDR.LATVKAHPKYLN.IAIS.FFLAAKTVEEDEKIPVLKVLARDSFCGCSSSEILRMERIILDKLN | |
| Retrocopy | SSLLDRILATVKAHPKYLNYIAISWFFLAAKTVEEDEKIPVLKVLARDSFCGCSSSEILRMERIILDKLN |
| Parental | WDLHTATPLDFLHIFHAIAVSARPQLLFSLPKLSPSQHLAVLTKQLLHCMACNQLLQFKGSMLALAMVSL |
| WDLH.ATPLDFLHIFHAI.VS.RPQLLFS.PKLSPSQHL..LTK.LL.CMACNQLLQFKGSMLALAMVSL | |
| Retrocopy | WDLHMATPLDFLHIFHAIVVSTRPQLLFSWPKLSPSQHLVILTKKLLPCMACNQLLQFKGSMLALAMVSL |
| Parental | EMEKLIPDWLPLTIELLQKAQMDSSQLIHCR-ELVAYHLSALQSALPLNSVYVYRPLKHTLVTCDKGAFK |
| EMEK.IPDWLPLTIELLQ.A.M.SSQ.IHCR.ELVAYH.SA.QSALPLNSVYVYRPLKHTLVTCDKGAF. | |
| Retrocopy | EMEKPIPDWLPLTIELLQEAEMGSSQ*IHCR>ELVAYHCSAPQSALPLNSVYVYRPLKHTLVTCDKGAFT |
| Parental | LHPSSVSGPDFSKDNSKPEVPVRGPAAFHLHLPAASGCKQTSAKRKVEEMEVDDFYDGIKRLYNEDNGPE |
| LHPSSVSGPDFSKD.SKPEVPVRGPAAFHLHLPAASGCKQTSAK.KVEEMEVDDF..GIK.LYNEDNGPE | |
| Retrocopy | LHPSSVSGPDFSKDKSKPEVPVRGPAAFHLHLPAASGCKQTSAKWKVEEMEVDDFSNGIKPLYNEDNGPE |
| Parental | NVGSVCGTDLSRQEGHASPCPPLQPVSVM |
| NVGSVCGTDLSRQEGHASPCPPLQPVSVM | |
| Retrocopy | NVGSVCGTDLSRQEGHASPCPPLQPVSVM |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .05 RPM | 150 .36 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 150 .41 RPM |
| SRP007412_heart | 0 .06 RPM | 76 .42 RPM |
| SRP007412_kidney | 0 .12 RPM | 174 .31 RPM |
| SRP007412_liver | 0 .00 RPM | 83 .07 RPM |
| SRP007412_testis | 1 .28 RPM | 161 .18 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_2730 was not detected
No EST(s) were mapped for retro_mmus_2730 retrocopy.
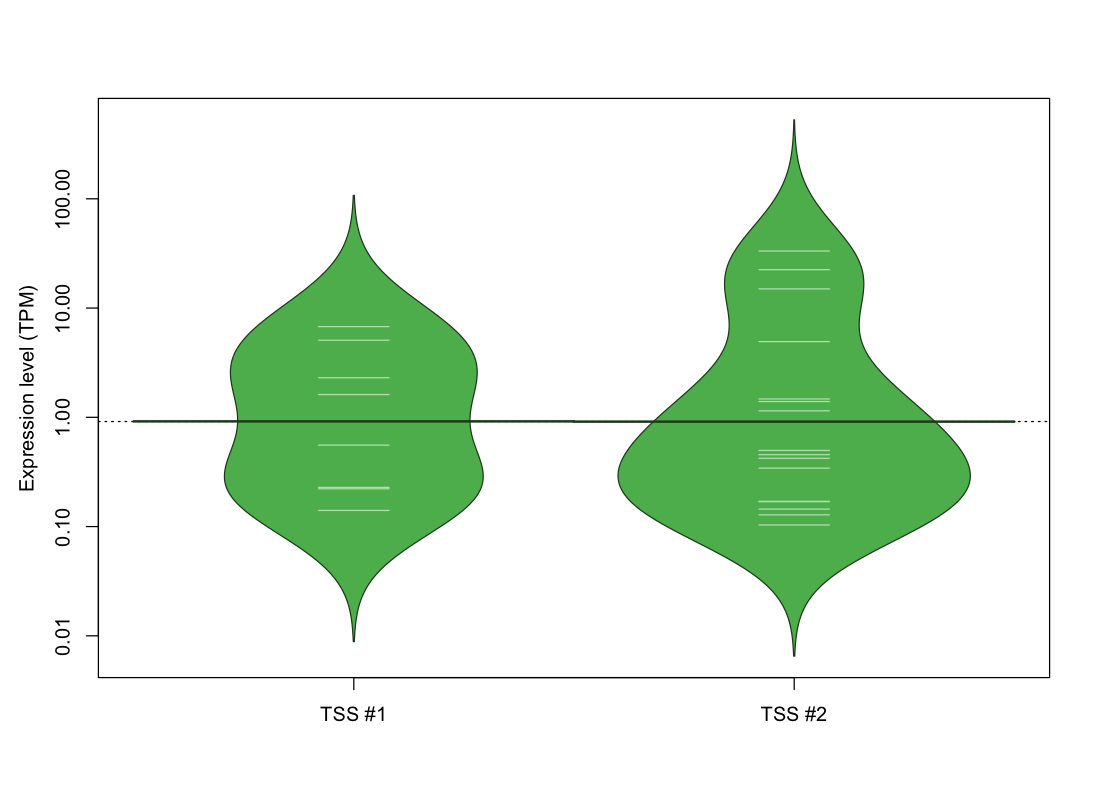
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_122524 | 1064 libraries | 4 libraries | 2 libraries | 2 libraries | 0 libraries |
| TSS #2 | TSS_122525 | 1056 libraries | 9 libraries | 4 libraries | 0 libraries | 3 libraries |
The graphical summary, for retro_mmus_2730 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2730 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2730 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 8 parental genes, and 9 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000014768 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000000952 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000029218 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000019904 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000020326 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000063015 | 2 retrocopies |
retro_mmus_1211, retro_mmus_2730 ,
|
| Tarsius syrichta | ENSTSYG00000009907 | 1 retrocopy | |
| Vicugna pacos | ENSVPAG00000011865 | 1 retrocopy |