RetrogeneDB ID: | retro_mmus_30 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:23101769..23102231(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000073730 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ppp1r14b | ||
| Ensembl ID: | ENSMUSG00000056612 | ||
| Aliases: | Ppp1r14b, AOM172, PHI-1, PLCB3N, Png | ||
| Description: | protein phosphatase 1, regulatory (inhibitor) subunit 14B [Source:MGI Symbol;Acc:MGI:107682] |

Retrocopy-Parental alignment summary:
>retro_mmus_30
ATGGCGGACAGCGGCCCCTCGGGGGGGCCAGCTTTGACTGATCCAGGCCCCGAGCCAGGCAGTGCCAACACAGGGCCCCG
AGTCTACTTCGAGAGTCCCCCTGGGACCCCAGGCGAAACCCCTGGCCCCGGGAACAACCCGGATAAAGATGACCAGGTGA
GACGGCCACATAGGCCAGATAAGATCATCGTCAAGTACGACCGCAAGGAGCTACGGAAGCGCCTCAACCTGGACGACTGG
ATCTTAGAACAGCTCACTGGTCTCTACGACTGCAAGGAAGAGGAAATCCCAGAGCTCGAGATCGATGTGGACGAACTCTT
AGACATGGAAAGCGACGATACCCGGGCTGCTAGAGTCAAGGAGCTGTTAGTTGACTGTTACAAACCCACTGAGGCCTTCA
TCAATGACCTGCTAGACAGGATCCGGGGCATGCAGAAGCTGACCATACCCCTGAAGAACTAA
ATGGCGGACAGCGGCCCCTCGGGGGGGCCAGCTTTGACTGATCCAGGCCCCGAGCCAGGCAGTGCCAACACAGGGCCCCG
AGTCTACTTCGAGAGTCCCCCTGGGACCCCAGGCGAAACCCCTGGCCCCGGGAACAACCCGGATAAAGATGACCAGGTGA
GACGGCCACATAGGCCAGATAAGATCATCGTCAAGTACGACCGCAAGGAGCTACGGAAGCGCCTCAACCTGGACGACTGG
ATCTTAGAACAGCTCACTGGTCTCTACGACTGCAAGGAAGAGGAAATCCCAGAGCTCGAGATCGATGTGGACGAACTCTT
AGACATGGAAAGCGACGATACCCGGGCTGCTAGAGTCAAGGAGCTGTTAGTTGACTGTTACAAACCCACTGAGGCCTTCA
TCAATGACCTGCTAGACAGGATCCGGGGCATGCAGAAGCTGACCATACCCCTGAAGAACTAA
ORF - retro_mmus_30 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 75.66 % |
| Parental protein coverage: | 99.32 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MADSGPAGGAALAAPAPGPGSGSTGPRVYFQSPPGAAGEGPG---GADDDGPVR---RQGKVTVKYDRKE |
| MADSGP.GG.AL..P.P.PGS..TGPRVYF.SPPG..GE.PG.....D.D..VR...R..K..VKYDRKE | |
| Retrocopy | MADSGPSGGPALTDPGPEPGSANTGPRVYFESPPGTPGETPGPGNNPDKDDQVRRPHRPDKIIVKYDRKE |
| Parental | LRKRLNLEEWILEQLTRLYDCQEEEIPELEIDVDELLDMESDDTRAARVKELLVDCYKPTEAFISGLLDK |
| LRKRLNL..WILEQLT.LYDC.EEEIPELEIDVDELLDMESDDTRAARVKELLVDCYKPTEAFI..LLD. | |
| Retrocopy | LRKRLNLDDWILEQLTGLYDCKEEEIPELEIDVDELLDMESDDTRAARVKELLVDCYKPTEAFINDLLDR |
| Parental | IRGMQKLSTPQK |
| IRGMQKL..P.K | |
| Retrocopy | IRGMQKLTIPLK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .02 RPM | 4 .72 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 4 .43 RPM |
| SRP007412_heart | 0 .00 RPM | 11 .52 RPM |
| SRP007412_kidney | 0 .00 RPM | 9 .76 RPM |
| SRP007412_liver | 0 .03 RPM | 6 .36 RPM |
| SRP007412_testis | 78 .99 RPM | 27 .86 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_30 was not detected
No EST(s) were mapped for retro_mmus_30 retrocopy.
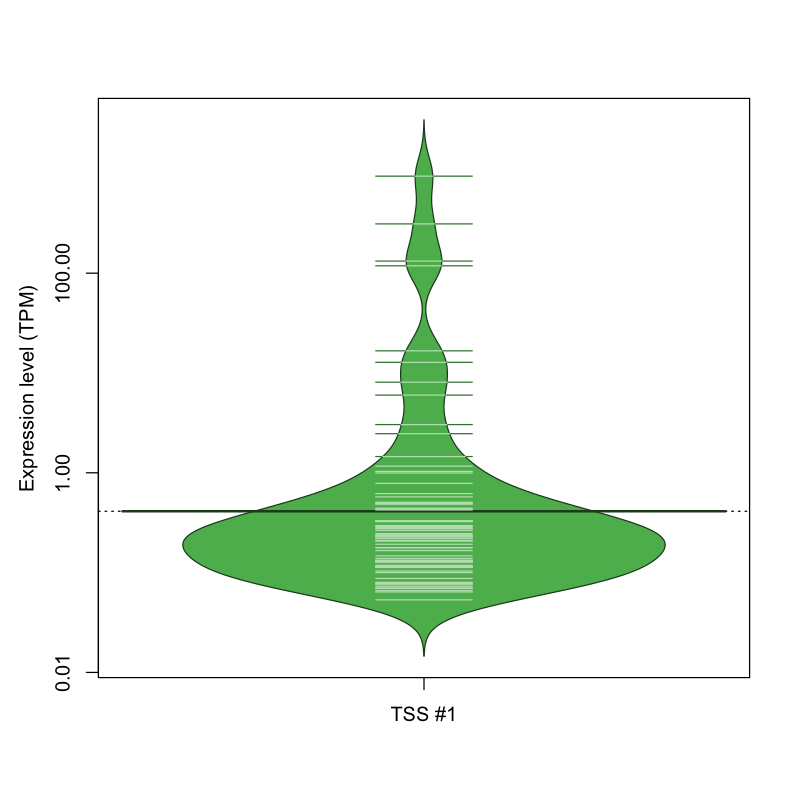
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_73669 | 1000 libraries | 59 libraries | 5 libraries | 2 libraries | 6 libraries |
The graphical summary, for retro_mmus_30 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_30 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_30 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_41 |
Parental genes homology:
Parental genes homology involve 7 parental genes, and 14 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000018019 | 1 retrocopy | |
| Felis catus | ENSFCAG00000030667 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000040653 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000056612 | 5 retrocopies | |
| Otolemur garnettii | ENSOGAG00000030926 | 2 retrocopies | |
| Pteropus vampyrus | ENSPVAG00000017120 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000021151 | 2 retrocopies |