RetrogeneDB ID: | retro_mmus_233 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 11:22973227..22974514(+) | ||
| Located in intron of: | ENSMUSG00000051355 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000044068 | |
| Aliases: | Zrsr1, D11Ncvs75, Irlgs2, SP2, U2af1-rs1, U2afbp-rs | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Zrsr2 | ||
| Ensembl ID: | ENSMUSG00000031370 | ||
| Aliases: | Zrsr2, 35kDa, 5031411E02Rik, A230052C13Rik, C77286, U2af1-rs2, URP | ||
| Description: | zinc finger (CCCH type), RNA binding motif and serine/arginine rich 2 [Source:MGI Symbol;Acc:MGI:103287] |

Retrocopy-Parental alignment summary:
>retro_mmus_233
ATGGCATCACGGCAGACCGCGATTCCTGAGAAACTCAGCCGAAAACAATACAAGGCGGCAATGAAGAAGGAGAAACGCAA
GAAACGTCGGCAGAAAATGGCTCGGCTGAGAGCTCTGGAAGCCCCACCAGAGGAGGACGATGATGTTTCTGCTAACGAAG
AACTTGCAGAGCGATTACTGGAGATAGAGCGGCAAAGATTACATGAAGAGTGGCTGCTGAGGGAGGAGAAGGCGCAAGAA
GAATTCAGAATAAAGAAGAAAAAGGAAGAGGCCGCTAGAAAACAGAAGGAAGAACAGGAGAGACAAATAAAGGCCGAGTG
GGAAGAACAACAGAAAAAACAGAGAGAGGAGGAGGAGCAGAAGCTACAGGAGAAGAGAGAGAGGGAGGAAGCGGTGCAGA
AAATGCTGGACCAGGCTGAAAATGAGCGCATTTGGCAGAACCCGGAACCACCCAAGGATTTAAGGCTGGAGAAATATCGA
CCCAGTTGTCCCTTCTACAATAAAACGGGAGCGTGCAGATTTGGTAACAGATGTTCACGGAAACACGACTTTCCCACGTC
AAGTCCCACCCTTCTTGTGAAGAGTATGTTTACAACGTTTGGAATGGAGCAGTGCAGAAGGGATGACTATGACTCAGACG
CAAACCTGGAGTACAGTGAGGAGGAGACCTACCAGCAGTTCTTGGATTTCTACCATGACGTGCTGCCGGAGTTCAAGAAC
GTGGGAAAGGTGATTCAGTTCAAAGTAAGCTGCAACCTGGAACCTCATCTGCGGGGCAATGTGTATGTTCAGTACCAGTC
GGAAGAAGAATGCCAAGCAGCCCTCTCTCTCTTTAATGGAAGATGGTACGCAGGACGACAGCTCCAGTGTGAATTCTGTC
CAGTGACCCGGTGGAAGGTTGCAATTTGTGGTTTATTCGAAATGCAAAAGTGTCCAAAAGGAAAGCACTGCAACTTCCTT
CATGTGTTCAGAAATCCCAACAACGAATTTAGAGAAGCTAACAGAGACATCTACATGTCTCCTCCGGCTTGGACTGGCTC
CTCTGGTAAAAACTCAGACAGGAGGGAAAGGAAGGACCATCATGAGGAATACTATAGCAAGTCAAGAAGCTACCACTCTG
GTTCATACCACTCCTCCAAGAGAAACAGGGAGTCCGAGAGGAAGAGTCCTCACAGGTGGAAGAAATCTCACAAACAGACA
ACGAAGAGTCATGAGAGGCACAGTTCAAGAAGAGGAAGAGAAGAGGACAGCAGTCCAGGTCCACAAAGCCAGAGCCACAG
AACCTGA
ORF - retro_mmus_233 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 73.73 % |
| Parental protein coverage: | 80.22 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | TAGPQKLSRKKYLALRKKERRKRRRQALARLREAELAQKEEEEDPLAEEKRLEEERLLEEERQRLHEEWL |
| TA.P.KLSRK.Y.A..KKE.RK.RRQ..ARLR..E.A..EE..D..A.E...E..RLLE.ERQRLHEEWL | |
| Retrocopy | TAIPEKLSRKQYKAAMKKEKRKKRRQKMARLRALE-APPEEDDDVSANEELAE--RLLEIERQRLHEEWL |
| Parental | LREEKAQEEFRAKKKKEEEARKRKEELERKLKAEWEEQQRKEREEEEQKRQEKREREEAVQKMLDQAENE |
| LREEKAQEEFR.KKKKEE.ARK.KEE.ER..KAEWEEQQ.K.REEEEQK.QEKREREEAVQKMLDQA... | |
| Retrocopy | LREEKAQEEFRIKKKKEEAARKQKEEQERQIKAEWEEQQKKQREEEEQKLQEKREREEAVQKMLDQA--- |
| Parental | LENGGTWQNPEPPMDIRVLEKDRANCPFYSKTGACRFGDRCSRKHNFPTSSPTLLIKGMFTTFGMEQCRR |
| .EN...WQNPEPP.D.R.LEK.R..CPFY.KTGACRFG.RCSRKH.FPTSSPTLL.K.MFTTFGMEQCRR | |
| Retrocopy | -ENERIWQNPEPPKDLR-LEKYRPSCPFYNKTGACRFGNRCSRKHDFPTSSPTLLVKSMFTTFGMEQCRR |
| Parental | DDYDPDSSLEFSEEEIYQQFLDFYYDVLPEFKSVGKVIQFKVSCNLEPHLRGNVYVQYQSEEDCQAAFSV |
| DDYD.D..LE.SEEE.YQQFLDFY.DVLPEFK.VGKVIQFKVSCNLEPHLRGNVYVQYQSEE.CQAA.S. | |
| Retrocopy | DDYDSDANLEYSEEETYQQFLDFYHDVLPEFKNVGKVIQFKVSCNLEPHLRGNVYVQYQSEEECQAALSL |
| Parental | FNGRWYAGRQLQCEFCPVTRWKMAICGLFEVQQCPRGKHCNFLHVFRNPNNEYRDANRDLYPSPDWTSSS |
| FNGRWYAGRQLQCEFCPVTRWK.AICGLFE.Q.CP.GKHCNFLHVFRNPNNE.R.ANRD.Y.SP.....S | |
| Retrocopy | FNGRWYAGRQLQCEFCPVTRWKVAICGLFEMQKCPKGKHCNFLHVFRNPNNEFREANRDIYMSPPAWTGS |
| Parental | FGKNSERRERASHYDEYYGRSRRRRRSPSPDFYKRNGESDRKSSSRHRVKKSHRYGMKSRERRSSPSRRR |
| .GKNS.RRER..H..EYY..S.R...S.S....KRN.ES.RKS...HR.KKSH....KS.ER.SS...R. | |
| Retrocopy | SGKNSDRRERKDHHEEYYSKS-RSYHSGSYHSSKRNRESERKSP--HRWKKSHKQTTKSHERHSSRRGRE |
| Parental | KDHSPGPWSQSRRS |
| .D.SPGP.SQS.R. | |
| Retrocopy | EDSSPGPQSQSHRT |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 46 .68 RPM | 35 .39 RPM |
| SRP007412_cerebellum | 84 .24 RPM | 34 .09 RPM |
| SRP007412_heart | 18 .09 RPM | 13 .54 RPM |
| SRP007412_kidney | 29 .45 RPM | 25 .12 RPM |
| SRP007412_liver | 22 .07 RPM | 9 .59 RPM |
| SRP007412_testis | 7 .91 RPM | 6 .63 RPM |
RNA Polymerase II activity may be related with retro_mmus_233 in 4 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 11:22971831..22973134 |
| ENCFF001YIJ | POLR2A | 11:22972725..22973448 |
| ENCFF001YJZ | POLR2A | 11:22971653..22974036 |
| ENCFF001YKA | POLR2A | 11:22971681..22973971 |
6 EST(s) were mapped to retro_mmus_233 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AV574470 | 22973453 | 22973954 | 99.6 | 498 | 2 | 495 |
| BB618009 | 22973218 | 22973870 | 99.4 | 646 | 4 | 642 |
| BE656540 | 22973567 | 22973964 | 100 | 397 | 0 | 397 |
| BI409173 | 22973195 | 22973425 | 98.3 | 230 | 0 | 229 |
| BP776891 | 22973203 | 22973425 | 99.6 | 221 | 1 | 220 |
| BY643991 | 22973205 | 22973485 | 97.2 | 275 | 3 | 271 |
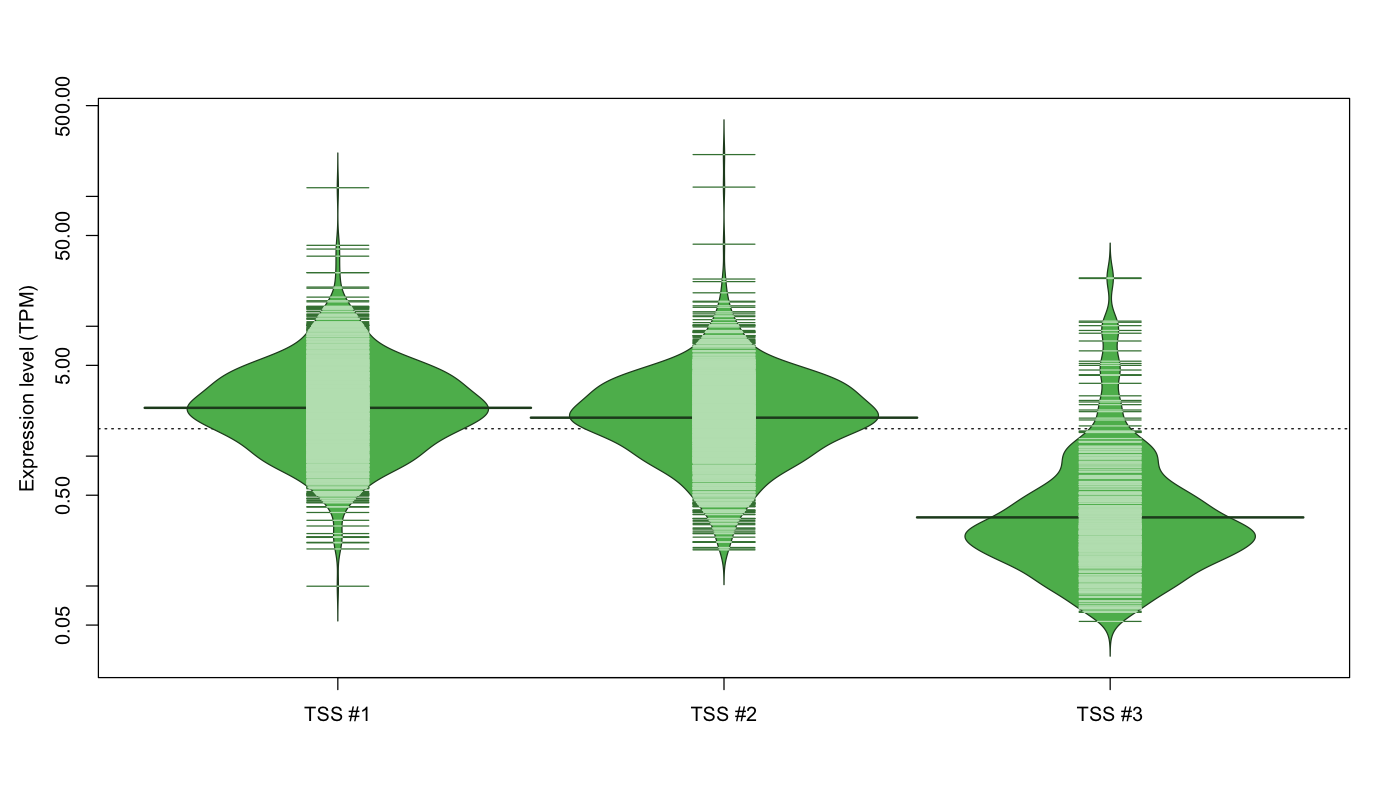
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_11571 | 78 libraries | 143 libraries | 694 libraries | 118 libraries | 39 libraries |
| TSS #2 | TSS_11572 | 127 libraries | 182 libraries | 674 libraries | 69 libraries | 20 libraries |
| TSS #3 | TSS_11573 | 719 libraries | 304 libraries | 38 libraries | 6 libraries | 5 libraries |
The graphical summary, for retro_mmus_233 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_233 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_233 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_176 |
Parental genes homology:
Parental genes homology involve 11 parental genes, and 13 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Felis catus | ENSFCAG00000009333 | 1 retrocopy | |
| Homo sapiens | ENSG00000169249 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000011962 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000014054 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000031370 | 1 retrocopy |
retro_mmus_233 ,
|
| Mus musculus | ENSMUSG00000061613 | 3 retrocopies | |
| Mus musculus | ENSMUSG00000078765 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000009496 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000021693 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000033176 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000005278 | 1 retrocopy |