RetrogeneDB ID: | retro_mmus_83 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | X:153498453..153500370(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000050148 | |
| Aliases: | Ubqln2, Chap1, Dsk2, HRIHFB2157, Plic-2, Plic2 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ubqln1 | ||
| Ensembl ID: | ENSMUSG00000005312 | ||
| Aliases: | Ubqln1, 1110046H03Rik, 1810030E05Rik, AU019746, C77538, D13Ertd372e, Da41, Dsk2, Plic-1, Plic1, Xdrp1 | ||
| Description: | ubiquilin 1 [Source:MGI Symbol;Acc:MGI:1860276] |

Retrocopy-Parental alignment summary:
>retro_mmus_83
ATGGCTGAGAACGGCGAGAGCAGCGGCCCCCCGCGCCCCTCCCGCGGCCCTGCGGCGGCCCCAGGCGCGGCCAGCCCGCC
GGCCGAGCCCAAAATCATCAAAGTCACTGTGAAGACCCCCAAAGAGAAGGAGGAGTTCGCGGTGCCCGAGAACAGCACCG
TGCAGCAGTTCAAGGAAGCGATTTCCAAACGCTTCAAATCGCAGACCGATCAGCTCGTGCTGATTTTCGCCGGAAAGATC
CTGAAAGATCAAGATACCTTGATGCAGCATGGCATCCATGATGGACTGACTGTTCACCTGGTTATCAAAAGCCAGAACCG
TCCGCAGGGCCAGGCCACCACGCAGCCCAGCACGACTGCGGGAACGAGCACGACGACCACGACCACCACCACGGCGGCGG
CGCCGGCGGCGACGACATCATCGGCTCCCAGGAGTAGCTCCACACCTACCACCACGAATAGCAGCTCCTTTGGGCTGGGA
AGCCTTTCCAGCCTTAGTAACCTGGGCTTGAACTCGCCCAACTTCACCGAGCTTCAGAACCAGATGCAGCAGCAGCTCCT
GGCCAGCCCTGAGATGATGATCCAGATCATGGAAAATCCCTTTGTTCAGAGCATGCTTTCGAATCCTGATCTGATGAGGC
AGCTCATCATGGCCAATCCACAGATGCAACAATTGATCCAGAGAAACCCAGAAATCAGCCACCTACTGAACAACCCAGAT
ATAATGAGGCAGACGCTGGAAATCGCCAGGAATCCTGCCATGATGCAAGAGATGATGCGAAATCAAGACCTGGCTCTCAG
TAATCTCGAAAGCATCCCAGGTGGCTACAATGCTCTGCGGCGCATGTACACTGACATTCAAGAACCTATGCTGAATGCCG
CACAGGAGCAGTTTGGGGGCAACCCGTTTGCCACGGTGGGGAGCAGTTCCACCTCAGGGGAAGGTACTCAGCCCTCCCGC
ACGGAGAATCGTGATCCGCTGCCCAATCCTTGGGCACCACCGCCAACAACCCAGACGGCTGCGACCACCACCACCACCAC
GACCACAAGCAGTGGCAGTGGCTCTGGCAGCAGCTCCAGCAGCACCACCGCCGGGAACACCATGGCTGCAGCTAATTACG
TGGCTAGCATCTTCAGCACCCCAGGAATGCAGAGCCTGCTGCAGCAGATAACTGAGAATCCCCAGCTGATTCAGAATATG
CTGTCTGCGCCCTACATGCGGAGCATGATGCAGTCGCTGAGCCAGAATCCAGATATGGCGGCCCAGATGATGCTGAGTAG
CCCGCTGTTCACTTCGAATCCTCAGCTGCAGGAGCAGATGCGTCCACAGCTCCCGAATTTCCTGCAGCAGATGCAGAATC
CAGAAACCATCGCGGCCATGTCAAACCCGAGAGCGATGCAAGCGCTAATGCAGATCCAGCAGGGGCTACAGACTCTAGCC
ACTGAAGCACCTGGCCTCATTCCCAGCTTCGCTCCAGGTGTGGGGATGGGAGTGCTGGGAACCGCCATAACCCCTGTGGG
CCCAGTCACTCCCATAGGGCCCATCGGCCCTATAGTTCCTTTCACCCCCATAGGCCCCATCGGGCCTATAGGACCCACTG
GCCCTGCAAGCTCCCCCGGCTCCACCGGCACTGGCATCCCGCCTGCAACCACTGTGTCCAGCTCTGCACCTACTGAAACC
ATAAGTCCAACCTCGGAATCTGGACCCAACCAGCAGTTCATTCAACAAATGGTGCAGGCTCTCACTGGAGGAAGCCCTCC
ACAGCCGCCGAATCCTGAAGTGAGATTTCAGCAGCAACTGGAACAGCTCAACGCCATGGGGTTCTTAAACCGCGAAGCAA
ACTTGCAGGCCCTAATAGCAACAGGAGGCGACATCAACGCGGCCATTGAGAGGCTGCTGGGCTCTCAGCCATCCTAA
ATGGCTGAGAACGGCGAGAGCAGCGGCCCCCCGCGCCCCTCCCGCGGCCCTGCGGCGGCCCCAGGCGCGGCCAGCCCGCC
GGCCGAGCCCAAAATCATCAAAGTCACTGTGAAGACCCCCAAAGAGAAGGAGGAGTTCGCGGTGCCCGAGAACAGCACCG
TGCAGCAGTTCAAGGAAGCGATTTCCAAACGCTTCAAATCGCAGACCGATCAGCTCGTGCTGATTTTCGCCGGAAAGATC
CTGAAAGATCAAGATACCTTGATGCAGCATGGCATCCATGATGGACTGACTGTTCACCTGGTTATCAAAAGCCAGAACCG
TCCGCAGGGCCAGGCCACCACGCAGCCCAGCACGACTGCGGGAACGAGCACGACGACCACGACCACCACCACGGCGGCGG
CGCCGGCGGCGACGACATCATCGGCTCCCAGGAGTAGCTCCACACCTACCACCACGAATAGCAGCTCCTTTGGGCTGGGA
AGCCTTTCCAGCCTTAGTAACCTGGGCTTGAACTCGCCCAACTTCACCGAGCTTCAGAACCAGATGCAGCAGCAGCTCCT
GGCCAGCCCTGAGATGATGATCCAGATCATGGAAAATCCCTTTGTTCAGAGCATGCTTTCGAATCCTGATCTGATGAGGC
AGCTCATCATGGCCAATCCACAGATGCAACAATTGATCCAGAGAAACCCAGAAATCAGCCACCTACTGAACAACCCAGAT
ATAATGAGGCAGACGCTGGAAATCGCCAGGAATCCTGCCATGATGCAAGAGATGATGCGAAATCAAGACCTGGCTCTCAG
TAATCTCGAAAGCATCCCAGGTGGCTACAATGCTCTGCGGCGCATGTACACTGACATTCAAGAACCTATGCTGAATGCCG
CACAGGAGCAGTTTGGGGGCAACCCGTTTGCCACGGTGGGGAGCAGTTCCACCTCAGGGGAAGGTACTCAGCCCTCCCGC
ACGGAGAATCGTGATCCGCTGCCCAATCCTTGGGCACCACCGCCAACAACCCAGACGGCTGCGACCACCACCACCACCAC
GACCACAAGCAGTGGCAGTGGCTCTGGCAGCAGCTCCAGCAGCACCACCGCCGGGAACACCATGGCTGCAGCTAATTACG
TGGCTAGCATCTTCAGCACCCCAGGAATGCAGAGCCTGCTGCAGCAGATAACTGAGAATCCCCAGCTGATTCAGAATATG
CTGTCTGCGCCCTACATGCGGAGCATGATGCAGTCGCTGAGCCAGAATCCAGATATGGCGGCCCAGATGATGCTGAGTAG
CCCGCTGTTCACTTCGAATCCTCAGCTGCAGGAGCAGATGCGTCCACAGCTCCCGAATTTCCTGCAGCAGATGCAGAATC
CAGAAACCATCGCGGCCATGTCAAACCCGAGAGCGATGCAAGCGCTAATGCAGATCCAGCAGGGGCTACAGACTCTAGCC
ACTGAAGCACCTGGCCTCATTCCCAGCTTCGCTCCAGGTGTGGGGATGGGAGTGCTGGGAACCGCCATAACCCCTGTGGG
CCCAGTCACTCCCATAGGGCCCATCGGCCCTATAGTTCCTTTCACCCCCATAGGCCCCATCGGGCCTATAGGACCCACTG
GCCCTGCAAGCTCCCCCGGCTCCACCGGCACTGGCATCCCGCCTGCAACCACTGTGTCCAGCTCTGCACCTACTGAAACC
ATAAGTCCAACCTCGGAATCTGGACCCAACCAGCAGTTCATTCAACAAATGGTGCAGGCTCTCACTGGAGGAAGCCCTCC
ACAGCCGCCGAATCCTGAAGTGAGATTTCAGCAGCAACTGGAACAGCTCAACGCCATGGGGTTCTTAAACCGCGAAGCAA
ACTTGCAGGCCCTAATAGCAACAGGAGGCGACATCAACGCGGCCATTGAGAGGCTGCTGGGCTCTCAGCCATCCTAA
ORF - retro_mmus_83 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 73.71 % |
| Parental protein coverage: | 66.67 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | TNAPGSTVTSSPAPDSNPTSGSAANSSFGVGGLGGLAGLSSLGLNTTNFSELQSQMQRQLLSNPEMMVQI |
| T.A.....T.S.AP.S..T......SSF...GLG.L..LS.LGLN..NF.ELQ.QMQ.QLL..PEMM.QI | |
| Retrocopy | TTAAAPAATTSSAPRSSSTPTTTNSSSF---GLGSLSSLSNLGLNSPNFTELQNQMQQQLLASPEMMIQI |
| Parental | MENPFVQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHMLNNPDIMRQTLELARNPAMMQEMMRNQDRAL |
| MENPFVQSMLSNPDLMRQLIMANPQMQQLIQRNPEISH.LNNPDIMRQTLE.ARNPAMMQEMMRNQD.AL | |
| Retrocopy | MENPFVQSMLSNPDLMRQLIMANPQMQQLIQRNPEISHLLNNPDIMRQTLEIARNPAMMQEMMRNQDLAL |
| Parental | SNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFASLVSSSSSAEGTQPSRTENRDPLPNPWAPQTS |
| SNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFA...SSS.S.EGTQPSRTENRDPLPNPWAP... | |
| Retrocopy | SNLESIPGGYNALRRMYTDIQEPMLNAAQEQFGGNPFATVGSSSTSGEGTQPSRTENRDPLPNPWAPPPT |
| Parental | QSSPASGTTGSTTNTMSTSGGTATSTPAGQSTSGPSLVPGAGASMFNTPGMQSLLQQITENPQLMQNMLS |
| ....A..TT..TT...S.SG....ST.AG........V....AS.F.TPGMQSLLQQITENPQL.QNMLS | |
| Retrocopy | TQTAATTTTTTTTSSGSGSGSSSSSTTAGNTMAAANYV----ASIFSTPGMQSLLQQITENPQLIQNMLS |
| Parental | APYMRSMLQSLSQNPDLAAQMMLNNPLFAGNPQLQEQMRQQLPTFLQQMQNPDTLSAMSNPRAMQALLQI |
| APYMRSM.QSLSQNPD.AAQMML..PLF..NPQLQEQMR.QLP.FLQQMQNP.T..AMSNPRAMQAL.QI | |
| Retrocopy | APYMRSMMQSLSQNPDMAAQMMLSSPLFTSNPQLQEQMRPQLPNFLQQMQNPETIAAMSNPRAMQALMQI |
| Parental | QQGLQTLATEAPGLIPGFTPGLAAGNSGGSSGTNAPST |
| QQGLQTLATEAPGLIP.F.PG...G..G.......P.T | |
| Retrocopy | QQGLQTLATEAPGLIPSFAPGVGMGVLGTAITPVGPVT |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 124 .99 RPM | 168 .33 RPM |
| SRP007412_cerebellum | 106 .95 RPM | 186 .97 RPM |
| SRP007412_heart | 22 .54 RPM | 60 .70 RPM |
| SRP007412_kidney | 20 .89 RPM | 140 .12 RPM |
| SRP007412_liver | 10 .38 RPM | 150 .20 RPM |
| SRP007412_testis | 8 .64 RPM | 108 .22 RPM |
RNA Polymerase II actvity may be related with retro_mmus_83 in 3 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | X:153497811..153498499 |
| ENCFF001YJZ | POLR2A | X:153497245..153498777 |
| ENCFF001YKA | POLR2A | X:153497373..153498559 |
41 EST(s) were mapped to retro_mmus_83 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AA030192 | 153498672 | 153499107 | 99.4 | 432 | 2 | 428 |
| AA117575 | 153499751 | 153499923 | 98.9 | 170 | 2 | 168 |
| AI509123 | 153498677 | 153499107 | 98.9 | 425 | 3 | 418 |
| AI391077 | 153498491 | 153498899 | 99.8 | 407 | 1 | 406 |
| AI413170 | 153498472 | 153498865 | 100 | 393 | 0 | 393 |
| AI413921 | 153498472 | 153498931 | 99.4 | 459 | 0 | 458 |
| AI414229 | 153498444 | 153498783 | 100 | 339 | 0 | 339 |
| AI430809 | 153498444 | 153498756 | 99.4 | 310 | 2 | 308 |
| AV480727 | 153499416 | 153499929 | 99.9 | 510 | 1 | 507 |
| AW046593 | 153498445 | 153498835 | 96.2 | 378 | 12 | 365 |
| AW050079 | 153498445 | 153498800 | 100 | 355 | 0 | 355 |
| AW244687 | 153499180 | 153499479 | 99.7 | 298 | 1 | 297 |
| BE381327 | 153498739 | 153499361 | 98 | 618 | 2 | 609 |
| BE864671 | 153498979 | 153499431 | 99.4 | 448 | 3 | 445 |
| BE950186 | 153498445 | 153498924 | 99 | 477 | 2 | 474 |
| BE950766 | 153498445 | 153498913 | 99.2 | 464 | 4 | 460 |
| BE995880 | 153498444 | 153498752 | 100 | 308 | 0 | 308 |
| BF137260 | 153498870 | 153499653 | 98.5 | 721 | 5 | 699 |
| BG228726 | 153498480 | 153498938 | 99.4 | 458 | 0 | 457 |
| BG694578 | 153498915 | 153499367 | 98.1 | 443 | 9 | 434 |
| BM944608 | 153499135 | 153499744 | 99.9 | 606 | 1 | 603 |
| BQ715304 | 153498799 | 153499558 | 97.8 | 754 | 0 | 741 |
| BQ769737 | 153498591 | 153499292 | 100 | 697 | 0 | 696 |
| BU614196 | 153499186 | 153499768 | 100 | 582 | 0 | 582 |
| BU702010 | 153498885 | 153499598 | 100 | 713 | 0 | 713 |
| CA324121 | 153498512 | 153499250 | 100 | 737 | 0 | 737 |
| CA324233 | 153498410 | 153499219 | 100 | 809 | 0 | 809 |
| CA530125 | 153498844 | 153499444 | 99.9 | 599 | 1 | 598 |
| CB246615 | 153498517 | 153499250 | 100 | 732 | 0 | 732 |
| CD353970 | 153498660 | 153499335 | 100 | 673 | 0 | 673 |
| CD553142 | 153499090 | 153499209 | 100 | 119 | 0 | 119 |
| CF736553 | 153498645 | 153499353 | 100 | 705 | 0 | 705 |
| CN529501 | 153498571 | 153499385 | 99.8 | 803 | 1 | 800 |
| CN530225 | 153498571 | 153499199 | 100 | 628 | 0 | 628 |
| CV562253 | 153498784 | 153499442 | 100 | 658 | 0 | 658 |
| CX206222 | 153499374 | 153499918 | 99.3 | 543 | 1 | 541 |
| CX211713 | 153499111 | 153499672 | 99.7 | 559 | 2 | 557 |
| CX733159 | 153499083 | 153499725 | 99.9 | 641 | 1 | 640 |
| CX733792 | 153498948 | 153499575 | 99.9 | 626 | 1 | 625 |
| W30442 | 153498535 | 153498899 | 99.8 | 361 | 0 | 358 |
| W57195 | 153499435 | 153499830 | 100 | 395 | 0 | 395 |
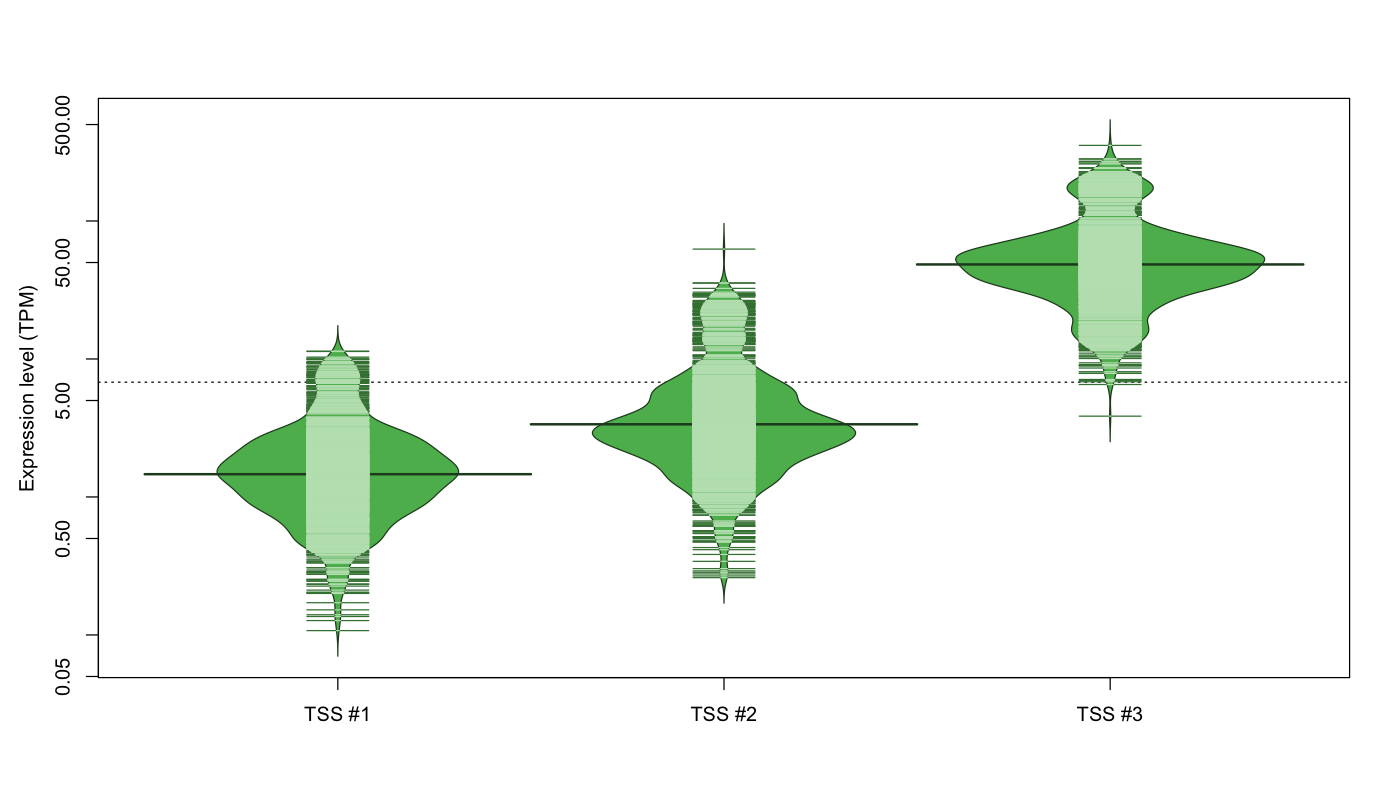
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_155947 | 163 libraries | 275 libraries | 570 libraries | 60 libraries | 4 libraries |
| TSS #2 | TSS_155948 | 69 libraries | 65 libraries | 658 libraries | 172 libraries | 108 libraries |
| TSS #3 | TSS_155949 | 3 libraries | 0 libraries | 1 library | 13 libraries | 1055 libraries |
The graphical summary, for retro_mmus_83 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_83 was not experimentally validated.