RetrogeneDB ID: | retro_hsap_2730 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 3:169649867..169651020(+) | ||
| Located in intron of: | ENSG00000187033 | ||
Retrocopyinformation | Ensembl ID: | ENSG00000244039 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | FAM20B | ||
| Ensembl ID: | ENSG00000116199 | ||
| Aliases: | FAM20B, gxk1 | ||
| Description: | family with sequence similarity 20, member B [Source:HGNC Symbol;Acc:23017] |

Retrocopy-Parental alignment summary:
>retro_hsap_2730
ATACATCAACTGCCAAGCAGCGTTTCACTACTTGATCCCCTGAGGGCTTTTCACCGAATGATGACTGGCTTGTGGATGGA
GCTGTCACCCAAGTTGGACCACACGTTGCAGTCTTCCTAGGAGACTGCAGCCCAGTGGGTGGTTCCCCAGGAAGTGTACT
GTGAAGAGACACCAGAGCTGGGGGCAATCATGCATGCCATGGCCACCAAGCAAATTATTAAAGCTGATGTGGGTTATAAA
GGGACACAGCTGAAAGCCTTACTGATCCTTGAAAGGGAACAGAAAGTTGTTTTCGAACCTAAGTAGTATAGCCGAGACTA
TGTAGTGGAAGGGAACCATATGCCGGTTATGATAGACACAATGCGGAGGTAGCAGCCTTTCACTTGGACAGGATTGTGGG
TGTCCCAGAGCCCTGCTGATGGTGGGCAGATCCGTTCATCTTCAGACAGAGATCAAGCCTGTTACCATGGAGCAGCTGTT
GAGCCCCTTCCTAACTGTAGGACACAATACTTGTTTTTATGGGAAGTGTTATTACTACCGAGAAACAGAATCAGCTTGTG
CCGATGGAGACACAATGGAAAAATCTGTCACACTTTGGCTTCCAGATGTGTGGCCTCTCCAGAAACACTGACACCCGTGG
GCCAGGACTTACTGAAAAGGCAAATTGGCCAGGTGGGAGTATGATGAGCGCTACTGTGAGGCTGTGAAGAAAACGTCCCT
TTATGACTCTGGCCTGTGCCGCCTGGACACCATTAACACAGCTGTCTTTGATTACCTGATTGACAATGCTGACTGCCATT
ACTATAAGAACTTTCAAGATAATGAGGGCGCCAGTATGCTCATCCTTCTTGATAACAACAAAAGCTTTGGGAACCCCTTG
CTGGATGAAAGAAGCATTCTTTCCCCCCCTCTATCAGTGTGGCATCACTCAGGTGTCTACCTGGAACAGACGGAACTACC
TAAAGAATGGTGTGCTGAAGTCTACCTTAAAATCTGCCATGGCCCATGACCCCCTCTCCCCAGTGCTCTCTGATCCTCGT
CTGGACGCCATGGACCAGTGGCTCCTGAGTGTCCTGGCCACCGTGGAGCAGTGCACTGACCAGTTTGGGATGGACACTGT
ATGGTAGAAGACACAATGCCTTTCTCCCACTTG
ATACATCAACTGCCAAGCAGCGTTTCACTACTTGATCCCCTGAGGGCTTTTCACCGAATGATGACTGGCTTGTGGATGGA
GCTGTCACCCAAGTTGGACCACACGTTGCAGTCTTCCTAGGAGACTGCAGCCCAGTGGGTGGTTCCCCAGGAAGTGTACT
GTGAAGAGACACCAGAGCTGGGGGCAATCATGCATGCCATGGCCACCAAGCAAATTATTAAAGCTGATGTGGGTTATAAA
GGGACACAGCTGAAAGCCTTACTGATCCTTGAAAGGGAACAGAAAGTTGTTTTCGAACCTAAGTAGTATAGCCGAGACTA
TGTAGTGGAAGGGAACCATATGCCGGTTATGATAGACACAATGCGGAGGTAGCAGCCTTTCACTTGGACAGGATTGTGGG
TGTCCCAGAGCCCTGCTGATGGTGGGCAGATCCGTTCATCTTCAGACAGAGATCAAGCCTGTTACCATGGAGCAGCTGTT
GAGCCCCTTCCTAACTGTAGGACACAATACTTGTTTTTATGGGAAGTGTTATTACTACCGAGAAACAGAATCAGCTTGTG
CCGATGGAGACACAATGGAAAAATCTGTCACACTTTGGCTTCCAGATGTGTGGCCTCTCCAGAAACACTGACACCCGTGG
GCCAGGACTTACTGAAAAGGCAAATTGGCCAGGTGGGAGTATGATGAGCGCTACTGTGAGGCTGTGAAGAAAACGTCCCT
TTATGACTCTGGCCTGTGCCGCCTGGACACCATTAACACAGCTGTCTTTGATTACCTGATTGACAATGCTGACTGCCATT
ACTATAAGAACTTTCAAGATAATGAGGGCGCCAGTATGCTCATCCTTCTTGATAACAACAAAAGCTTTGGGAACCCCTTG
CTGGATGAAAGAAGCATTCTTTCCCCCCCTCTATCAGTGTGGCATCACTCAGGTGTCTACCTGGAACAGACGGAACTACC
TAAAGAATGGTGTGCTGAAGTCTACCTTAAAATCTGCCATGGCCCATGACCCCCTCTCCCCAGTGCTCTCTGATCCTCGT
CTGGACGCCATGGACCAGTGGCTCCTGAGTGTCCTGGCCACCGTGGAGCAGTGCACTGACCAGTTTGGGATGGACACTGT
ATGGTAGAAGACACAATGCCTTTCTCCCACTTG
ORF - retro_hsap_2730 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 79.43 % |
| Parental protein coverage: | 94.13 % |
| Number of stop codons detected: | 4 |
| Number of frameshifts detected | 4 |
Retrocopy - Parental Gene Alignment:
| Parental | IDNLDTSAANREDQRAFHRMMTGLRVELAPKLDHTLQSPWEIAAQWVVPREVYPEETPELGAVMHAMATK |
| I..L..S.......RAFHRMMTGL..EL.PKLDHTLQS..E.AAQWVVP.EVY.EETPELGA.MHAMATK | |
| Retrocopy | IHQLPSSVSLLDPLRAFHRMMTGLWMELSPKLDHTLQSS*ETAAQWVVPQEVYCEETPELGAIMHAMATK |
| Parental | KIIKADVGYKGTQLKALLILEGGQKVVFKPKRYSRDHVVEG-EPYAGYDRHNAEVAAFHLDRILGFH-RA |
| .IIKADVGYKGTQLKALLILE..QKVVF.PK.YSRD.VVEG.EPYAGYDRHNAEVAAFHLDRI.G...RA | |
| Retrocopy | QIIKADVGYKGTQLKALLILEREQKVVFEPK*YSRDYVVEG<EPYAGYDRHNAEVAAFHLDRIVGVP<RA |
| Parental | PLVVGRFVNLRTEIKPVATEQLLSTFLTVGNNTCFYGKCYYCRETEPACADGDIMEGSVTLWLPDVWPLQ |
| .L.VGR.V.L.TEIKPV..EQLLS.FLTVG.NTCFYGKCYY.RETE.ACADGD.ME.SVTLWLPDVWPLQ | |
| Retrocopy | LLMVGRSVHLQTEIKPVTMEQLLSPFLTVGHNTCFYGKCYYYRETESACADGDTMEKSVTLWLPDVWPLQ |
| Parental | KHRHPWGRTYREGKLARWEYDESYCDAVKKTSPYDSGPRLLDIIDTAVFDYLIGNADRHHYESFQDDEGA |
| KH.HPW.RTY..GKLARWEYDE.YC.AVKKTS.YDSG...LD.I.TAVFDYLI.NAD.H.Y..FQD.EGA | |
| Retrocopy | KH*HPWARTY*KGKLARWEYDERYCEAVKKTSLYDSGLCRLDTINTAVFDYLIDNADCHYYKNFQDNEGA |
| Parental | SMLILLDNAKSFGNPSLDERSILA-PLYQCCIIRVSTWNRLNYLKNGVLKSALKSAMAHDPISPVLSDPH |
| SMLILLDN.KSFGNP.LDERSIL..PLYQC.I..VSTWNR.NYLKNGVLKS.LKSAMAHDP.SPVLSDP. | |
| Retrocopy | SMLILLDNNKSFGNPLLDERSILS>PLYQCGITQVSTWNRRNYLKNGVLKSTLKSAMAHDPLSPVLSDPR |
| Parental | LDAVDQRLLSVLATVKQCTDQFGMDTV-LVEDRMPLSHL |
| LDA.DQ.LLSVLATV.QCTDQFGMDTV..VED.MP.SHL | |
| Retrocopy | LDAMDQWLLSVLATVEQCTDQFGMDTV<MVEDTMPFSHL |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .00 RPM | 111 .02 RPM |
| bodymap2_adrenal | 0 .00 RPM | 108 .32 RPM |
| bodymap2_brain | 0 .00 RPM | 98 .26 RPM |
| bodymap2_breast | 0 .00 RPM | 129 .44 RPM |
| bodymap2_colon | 0 .00 RPM | 172 .36 RPM |
| bodymap2_heart | 0 .00 RPM | 99 .36 RPM |
| bodymap2_kidney | 0 .00 RPM | 86 .59 RPM |
| bodymap2_liver | 0 .00 RPM | 32 .83 RPM |
| bodymap2_lung | 0 .00 RPM | 89 .42 RPM |
| bodymap2_lymph_node | 0 .00 RPM | 67 .96 RPM |
| bodymap2_ovary | 0 .00 RPM | 105 .46 RPM |
| bodymap2_prostate | 0 .00 RPM | 132 .20 RPM |
| bodymap2_skeletal_muscle | 0 .00 RPM | 92 .74 RPM |
| bodymap2_testis | 0 .00 RPM | 93 .17 RPM |
| bodymap2_thyroid | 0 .00 RPM | 96 .27 RPM |
| bodymap2_white_blood_cells | 0 .00 RPM | 68 .95 RPM |
RNA Polymerase II actvity may be related with retro_hsap_2730 in 9 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CHO | POLR2A | 3:169650119..169650396 |
| ENCFF002CPG | POLR2A | 3:169649963..169650453 |
| ENCFF002CQA | POLR2A | 3:169649932..169650502 |
| ENCFF002CQC | POLR2A | 3:169649720..169650200 |
| ENCFF002CQC | POLR2A | 3:169649976..169650456 |
| ENCFF002CQG | POLR2A | 3:169649695..169650219 |
| ENCFF002CQG | POLR2A | 3:169649932..169650456 |
| ENCFF002CQK | POLR2A | 3:169649986..169650476 |
| ENCFF002CQM | POLR2A | 3:169649743..169650223 |
| ENCFF002CQM | POLR2A | 3:169649972..169650452 |
| ENCFF002CQO | POLR2A | 3:169649992..169650522 |
| ENCFF002CQO | POLR2A | 3:169649718..169650248 |
| ENCFF002CZY | POLR2A | 3:169649831..169650117 |
| ENCFF002CZY | POLR2A | 3:169650120..169650324 |
No EST(s) were mapped for retro_hsap_2730 retrocopy.
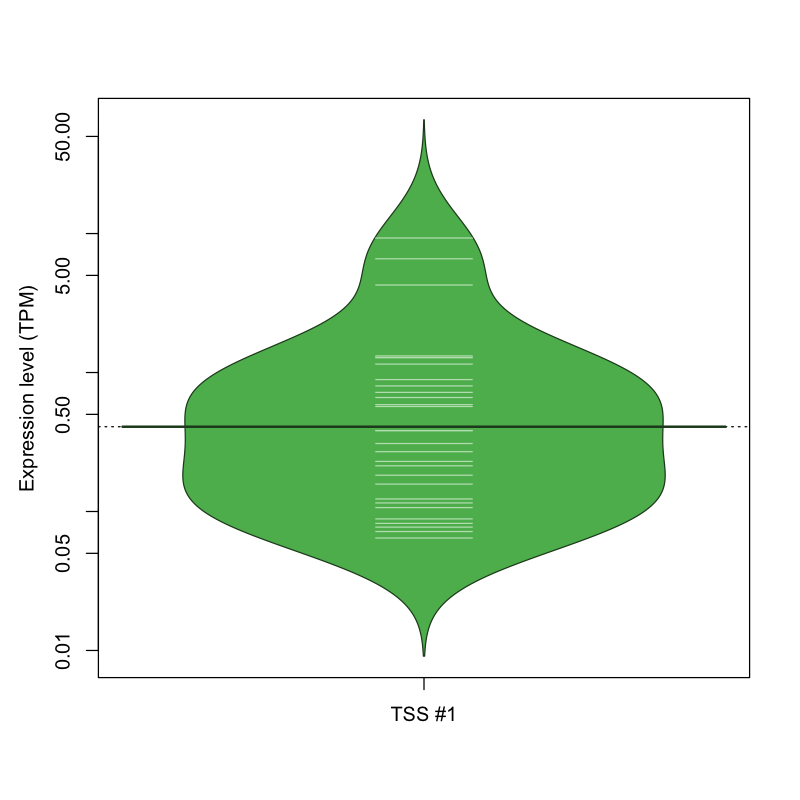
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_131526 | 1800 libraries | 22 libraries | 5 libraries | 2 libraries | 0 libraries |
The graphical summary, for retro_hsap_2730 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_2730 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_2730 has 2 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Pan troglodytes | retro_ptro_1845 |
| Pongo abelii | retro_pabe_2269 |
Parental genes homology:
Parental genes homology involve 13 parental genes, and 23 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Choloepus hoffmanni | ENSCHOG00000005236 | 6 retrocopies | |
| Dasypus novemcinctus | ENSDNOG00000006175 | 2 retrocopies | |
| Echinops telfairi | ENSETEG00000005651 | 1 retrocopy | |
| Homo sapiens | ENSG00000116199 | 1 retrocopy |
retro_hsap_2730 ,
|
| Gorilla gorilla | ENSGGOG00000002484 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000006654 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000006260 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000015580 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000012751 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000000466 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000001725 | 1 retrocopy | |
| Ictidomys tridecemlineatus | ENSSTOG00000005086 | 2 retrocopies | |
| Tarsius syrichta | ENSTSYG00000005482 | 3 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .11 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .26 RPM |
| CEU_NA12004 | 0 .30 RPM |
| CEU_NA12400 | 0 .04 RPM |
| CEU_NA12751 | 0 .05 RPM |
| CEU_NA12760 | 0 .04 RPM |
| CEU_NA12827 | 0 .05 RPM |
| CEU_NA12872 | 0 .03 RPM |
| CEU_NA12873 | 0 .13 RPM |
| FIN_HG00183 | 0 .05 RPM |
| FIN_HG00277 | 0 .15 RPM |
| FIN_HG00315 | 0 .28 RPM |
| FIN_HG00321 | 0 .03 RPM |
| FIN_HG00328 | 0 .14 RPM |
| FIN_HG00338 | 0 .28 RPM |
| FIN_HG00349 | 0 .17 RPM |
| FIN_HG00375 | 0 .12 RPM |
| FIN_HG00377 | 0 .18 RPM |
| FIN_HG00378 | 0 .21 RPM |
| GBR_HG00099 | 0 .17 RPM |
| GBR_HG00111 | 0 .09 RPM |
| GBR_HG00114 | 0 .18 RPM |
| GBR_HG00119 | 0 .22 RPM |
| GBR_HG00131 | 0 .23 RPM |
| GBR_HG00133 | 0 .12 RPM |
| GBR_HG00134 | 0 .15 RPM |
| GBR_HG00137 | 0 .19 RPM |
| GBR_HG00142 | 0 .22 RPM |
| GBR_HG00143 | 0 .32 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .12 RPM |
| TSI_NA20518 | 0 .25 RPM |
| TSI_NA20532 | 0 .10 RPM |
| TSI_NA20538 | 0 .32 RPM |
| TSI_NA20756 | 0 .03 RPM |
| TSI_NA20765 | 0 .35 RPM |
| TSI_NA20771 | 0 .17 RPM |
| TSI_NA20786 | 0 .26 RPM |
| TSI_NA20798 | 0 .22 RPM |
| YRI_NA18870 | 0 .10 RPM |
| YRI_NA18907 | 0 .10 RPM |
| YRI_NA18916 | 0 .08 RPM |
| YRI_NA19093 | 0 .21 RPM |
| YRI_NA19099 | 0 .13 RPM |
| YRI_NA19114 | 0 .21 RPM |
| YRI_NA19118 | 0 .25 RPM |
| YRI_NA19213 | 0 .07 RPM |
| YRI_NA19214 | 0 .15 RPM |
| YRI_NA19223 | 0 .22 RPM |