RetrogeneDB ID: | retro_hsap_459 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 1:158145639..158147520(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000227295 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | ELL2 | ||
| Ensembl ID: | ENSG00000118985 | ||
| Aliases: | None | ||
| Description: | elongation factor, RNA polymerase II, 2 [Source:HGNC Symbol;Acc:17064] |

Retrocopy-Parental alignment summary:
>retro_hsap_459
CAGCGCTATGGGCTCTCGTGCGGACGGCTGGGGCAGGACAACATCACCGTACTGCATGTGAAGCTCACCGAGTCGGCGAT
CCAGGCGCTCCAGACTTACCAGAGCCACAAGAATTTAATTTCTTTTCGACCTTCAATTCAGTTCCAAAGACTCCAGGAGC
TTGTCAAAATTCCCAAAAATGATCCCCTCAATGAAGTTCATAACTTTAACTTTTATTTGTCAAATGTGGGCAAAGACAAC
CCTCAGGGCAGCTTTGACTGCGTCCAGCAAACATTCTCCAGCTCTGGAGCCTCCCAGCTCAATTGCCTGGGATTTATACA
AGATAAAATTACAGTGTGTGCAACAAACGACTCGGATCAGATGACACGAGAAAGAACGACCCAGGCAGAGGAGGAATCCC
GCAACCGAAGCACAGAAGTTATCAAACCCGGTGGACCATATGTAGGGAAAAGAGTGCAAATTCGGAAAGCACCTCAAGCT
GTTTCAGATACAGTTCCTGAGAGGAAAAGGTCAACCCCCATGAACCCTGCAAATCCAATTCGATAGACACACAGCAGCAG
CAGCGTCTCTCAGAAGCCATACAGGGACAGGGTGGTTCACTTACTGGCCCTGAAGGCCTACAAGAAACCGGAGCTACTTG
CTAGACTCCAGAAAGATGGTGTCAATCAAAAAGACAGGAACTCCCTGGGAGCAATTCTGCAACAGGTAGCCAATCTGAAT
CCTAAGGACCTCTCGTATACTTCAAAGGATTATGTTTTTAAAGAGCTTCAGAGAGACTGGCCTGGATACAGTGAAATAGA
CACTTGGTCATTGGAGTCAATGCTCTCTAGAAAACTAAATCCATCTCAGAATGCTGCAGGCACCAGCCGTTCAGAATCTC
CAGTATGTTCTAGTAGAAACGCTGTATCTTCTCCTCAGACACGGCTTTTGGATTCAGAGTTTATTGATCATTTAATGGAT
AAAAAAGCCCGAATATCTCACCTGACGAGCAGAGTACCACCAACACTAAATGGTCATTTGAATCCCACCAGTGAAAAATC
TGCTGCAGGCCTCCCTCTGCCCCCTGCAGCTGCTGCCATCCCCACCCCTCCACCGCTTGCCTTCAACCCATCTGCCGGTT
TCACATCCTCCTCAGATTGTAAATTCTAACTCCAATTCCCCTAGCACTCCAGAAGGCCGGGGGCTCAAGACCTACCTGTT
GACAGTTTTAGTCAAAACAATAGTATCTATGAGAACCAACAAGACAAGTATACCTCTAGGACTTCTCTGGAAACCTTACC
CCCTGGTTCAGTTCTATTAAAGTGTCCAAAGCCTATGGAAGAAAACCATTCAGTGTCTCACAAAAAGTCCAAAAAGAAGT
CTAAAAAACATAAGGAAAAGGACCAAATAAAAAAGCACGACATTGAGACTATTGAGGAAAAGGAGAAAGATCTTAAGAGA
GAAGAGGAAATTGCCAAGCTAAATAACTCCAGTCCAAATTCCAGTGGAGGAGCTAAAGAGGATTGCACTGCCTCCATGGA
ACCTTCAGCAATTGAACTCCCAGATTATTTGATAAAATATATTGCTATCATCTCCTATGAGCAACGCCGGAATTATAAAG
ATGACGTCAGTGCAGAGTATGATGAGTACAGAGCTTTGCATGCCAGGATGGAGACTGTAGCTAGAAGATTTATCAAACTA
GATGCACAAAGAAAGCGCCTTTCTCCAGGCTCAAAAGAGTATCAGAATGTTCATGAAGAAGTCTTACAAGAATATCGGAA
GATAAAGCAGTCTAGTCCCAATTACCATGAAGAAAAATACAGATGTGAATATCTTCATAACAAGCTGGCTCACATCAAAA
GGCTACTAGGCGAATTTGACCAACAGCAGGCAGAGTCATGG
CAGCGCTATGGGCTCTCGTGCGGACGGCTGGGGCAGGACAACATCACCGTACTGCATGTGAAGCTCACCGAGTCGGCGAT
CCAGGCGCTCCAGACTTACCAGAGCCACAAGAATTTAATTTCTTTTCGACCTTCAATTCAGTTCCAAAGACTCCAGGAGC
TTGTCAAAATTCCCAAAAATGATCCCCTCAATGAAGTTCATAACTTTAACTTTTATTTGTCAAATGTGGGCAAAGACAAC
CCTCAGGGCAGCTTTGACTGCGTCCAGCAAACATTCTCCAGCTCTGGAGCCTCCCAGCTCAATTGCCTGGGATTTATACA
AGATAAAATTACAGTGTGTGCAACAAACGACTCGGATCAGATGACACGAGAAAGAACGACCCAGGCAGAGGAGGAATCCC
GCAACCGAAGCACAGAAGTTATCAAACCCGGTGGACCATATGTAGGGAAAAGAGTGCAAATTCGGAAAGCACCTCAAGCT
GTTTCAGATACAGTTCCTGAGAGGAAAAGGTCAACCCCCATGAACCCTGCAAATCCAATTCGATAGACACACAGCAGCAG
CAGCGTCTCTCAGAAGCCATACAGGGACAGGGTGGTTCACTTACTGGCCCTGAAGGCCTACAAGAAACCGGAGCTACTTG
CTAGACTCCAGAAAGATGGTGTCAATCAAAAAGACAGGAACTCCCTGGGAGCAATTCTGCAACAGGTAGCCAATCTGAAT
CCTAAGGACCTCTCGTATACTTCAAAGGATTATGTTTTTAAAGAGCTTCAGAGAGACTGGCCTGGATACAGTGAAATAGA
CACTTGGTCATTGGAGTCAATGCTCTCTAGAAAACTAAATCCATCTCAGAATGCTGCAGGCACCAGCCGTTCAGAATCTC
CAGTATGTTCTAGTAGAAACGCTGTATCTTCTCCTCAGACACGGCTTTTGGATTCAGAGTTTATTGATCATTTAATGGAT
AAAAAAGCCCGAATATCTCACCTGACGAGCAGAGTACCACCAACACTAAATGGTCATTTGAATCCCACCAGTGAAAAATC
TGCTGCAGGCCTCCCTCTGCCCCCTGCAGCTGCTGCCATCCCCACCCCTCCACCGCTTGCCTTCAACCCATCTGCCGGTT
TCACATCCTCCTCAGATTGTAAATTCTAACTCCAATTCCCCTAGCACTCCAGAAGGCCGGGGGCTCAAGACCTACCTGTT
GACAGTTTTAGTCAAAACAATAGTATCTATGAGAACCAACAAGACAAGTATACCTCTAGGACTTCTCTGGAAACCTTACC
CCCTGGTTCAGTTCTATTAAAGTGTCCAAAGCCTATGGAAGAAAACCATTCAGTGTCTCACAAAAAGTCCAAAAAGAAGT
CTAAAAAACATAAGGAAAAGGACCAAATAAAAAAGCACGACATTGAGACTATTGAGGAAAAGGAGAAAGATCTTAAGAGA
GAAGAGGAAATTGCCAAGCTAAATAACTCCAGTCCAAATTCCAGTGGAGGAGCTAAAGAGGATTGCACTGCCTCCATGGA
ACCTTCAGCAATTGAACTCCCAGATTATTTGATAAAATATATTGCTATCATCTCCTATGAGCAACGCCGGAATTATAAAG
ATGACGTCAGTGCAGAGTATGATGAGTACAGAGCTTTGCATGCCAGGATGGAGACTGTAGCTAGAAGATTTATCAAACTA
GATGCACAAAGAAAGCGCCTTTCTCCAGGCTCAAAAGAGTATCAGAATGTTCATGAAGAAGTCTTACAAGAATATCGGAA
GATAAAGCAGTCTAGTCCCAATTACCATGAAGAAAAATACAGATGTGAATATCTTCATAACAAGCTGGCTCACATCAAAA
GGCTACTAGGCGAATTTGACCAACAGCAGGCAGAGTCATGG
ORF - retro_hsap_459 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 90.14 % |
| Parental protein coverage: | 97.97 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | QRYGLSCGRLGQDNITVLHVKLTETAIRALETYQSHKNLIPFRPSIQFQGLHGLVKIPKNDPLNEVHNFN |
| QRYGLSCGRLGQDNITVLHVKLTE.AI.AL.TYQSHKNLI.FRPSIQFQ.L..LVKIPKNDPLNEVHNFN | |
| Retrocopy | QRYGLSCGRLGQDNITVLHVKLTESAIQALQTYQSHKNLISFRPSIQFQRLQELVKIPKNDPLNEVHNFN |
| Parental | FYLSNVGKDNPQGSFDCIQQTFSSSGASQLNCLGFIQDKITVCATNDSYQMTRERMTQAEEESRNRSTKV |
| FYLSNVGKDNPQGSFDC.QQTFSSSGASQLNCLGFIQDKITVCATNDS.QMTRER.TQAEEESRNRST.V | |
| Retrocopy | FYLSNVGKDNPQGSFDCVQQTFSSSGASQLNCLGFIQDKITVCATNDSDQMTRERTTQAEEESRNRSTEV |
| Parental | IKPGGPYVGKRVQIRKAPQAVSDTVPERKRSTPMNPANTIRKTHSSSTISQRPYRDRVIHLLALKAYKKP |
| IKPGGPYVGKRVQIRKAPQAVSDTVPERKRSTPMNPAN.IR.THSSS..SQ.PYRDRV.HLLALKAYKKP | |
| Retrocopy | IKPGGPYVGKRVQIRKAPQAVSDTVPERKRSTPMNPANPIR*THSSSSVSQKPYRDRVVHLLALKAYKKP |
| Parental | ELLARLQKDGVNQKDKNSLGAILQQVANLNSKDLSYTLKDYVFKELQRDWPGYSEIDRRSLESVLSRKLN |
| ELLARLQKDGVNQKD.NSLGAILQQVANLN.KDLSYT.KDYVFKELQRDWPGYSEID..SLES.LSRKLN | |
| Retrocopy | ELLARLQKDGVNQKDRNSLGAILQQVANLNPKDLSYTSKDYVFKELQRDWPGYSEIDTWSLESMLSRKLN |
| Parental | PSQNAAGTSRSESPVCSSRDAVSSPQKRLLDSEFIDPLMNKKARISHLTNRVPPTLNGHLNPTSEKSAAG |
| PSQNAAGTSRSESPVCSSR.AVSSPQ.RLLDSEFID.LM.KKARISHLT.RVPPTLNGHLNPTSEKSAAG | |
| Retrocopy | PSQNAAGTSRSESPVCSSRNAVSSPQTRLLDSEFIDHLMDKKARISHLTSRVPPTLNGHLNPTSEKSAAG |
| Parental | LPLPPAAAAIPTPPP-LPSTYLPISHPPQIVNSNSNSPSTPEGR-GTQDLPVDSFSQNDSIYEDQQDKYT |
| LPLPPAAAAIPTPPP.LPST.LP.SHPPQIVNSNSNSPSTPEGR.G.QDLPVDSFSQN.SIYE.QQDKYT | |
| Retrocopy | LPLPPAAAAIPTPPP>LPSTHLPVSHPPQIVNSNSNSPSTPEGR<GAQDLPVDSFSQNNSIYENQQDKYT |
| Parental | SRTSLETLPPGSVLLKCPKPMEENHSMSHKKSKKKSKKHKEKDQIKKHDIETIEEKEEDLKREEEIAKLN |
| SRTSLETLPPGSVLLKCPKPMEENHS.SH..................HDIETIEEKE.DLKREEEIAKLN | |
| Retrocopy | SRTSLETLPPGSVLLKCPKPMEENHSVSHXXXXXXXXXXXXXXXXXXHDIETIEEKEKDLKREEEIAKLN |
| Parental | NSSPNSSGGVKEDCTASMEPSAIELPDYLIKYIAIVSYEQRQNYKDDFNAEYDEYRALHARMETVARRFI |
| NSSPNSSGG.KEDCTASMEPSAIELPDYLIKYIAI.SYEQR.NYKDD..AEYDEYRALHARMETVARRFI | |
| Retrocopy | NSSPNSSGGAKEDCTASMEPSAIELPDYLIKYIAIISYEQRRNYKDDVSAEYDEYRALHARMETVARRFI |
| Parental | KLDAQRKRLSPGSKEYQNVHEEVLQEYQKIKQSSPNYHEEKYRCEYLHNKLAHIKRLIGEFDQQQAESW |
| KLDAQRKRLSPGSKEYQNVHEEVLQEY.KIKQSSPNYHEEKYRCEYLHNKLAHIKRL.GEFDQQQAESW | |
| Retrocopy | KLDAQRKRLSPGSKEYQNVHEEVLQEYRKIKQSSPNYHEEKYRCEYLHNKLAHIKRLLGEFDQQQAESW |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .24 RPM | 155 .09 RPM |
| bodymap2_adrenal | 0 .00 RPM | 63 .33 RPM |
| bodymap2_brain | 0 .16 RPM | 114 .76 RPM |
| bodymap2_breast | 0 .02 RPM | 92 .28 RPM |
| bodymap2_colon | 0 .10 RPM | 66 .86 RPM |
| bodymap2_heart | 0 .02 RPM | 108 .35 RPM |
| bodymap2_kidney | 0 .25 RPM | 61 .98 RPM |
| bodymap2_liver | 0 .13 RPM | 112 .74 RPM |
| bodymap2_lung | 0 .19 RPM | 149 .98 RPM |
| bodymap2_lymph_node | 0 .18 RPM | 152 .90 RPM |
| bodymap2_ovary | 0 .00 RPM | 33 .37 RPM |
| bodymap2_prostate | 0 .03 RPM | 82 .20 RPM |
| bodymap2_skeletal_muscle | 0 .15 RPM | 91 .36 RPM |
| bodymap2_testis | 0 .53 RPM | 49 .70 RPM |
| bodymap2_thyroid | 0 .30 RPM | 87 .89 RPM |
| bodymap2_white_blood_cells | 1 .12 RPM | 17 .13 RPM |
RNA Polymerase II actvity may be related with retro_hsap_459 in 4 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CRO | POLR2A | 1:158147424..158147844 |
| ENCFF002DAH | POLR2A | 1:158147524..158147808 |
| ENCFF002DBO | POLR2A | 1:158147438..158147822 |
| ENCFF002DBP | POLR2A | 1:158147454..158147784 |
1 EST(s) were mapped to retro_hsap_459 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| BG928961 | 158145614 | 158145949 | 100 | 335 | 0 | 335 |
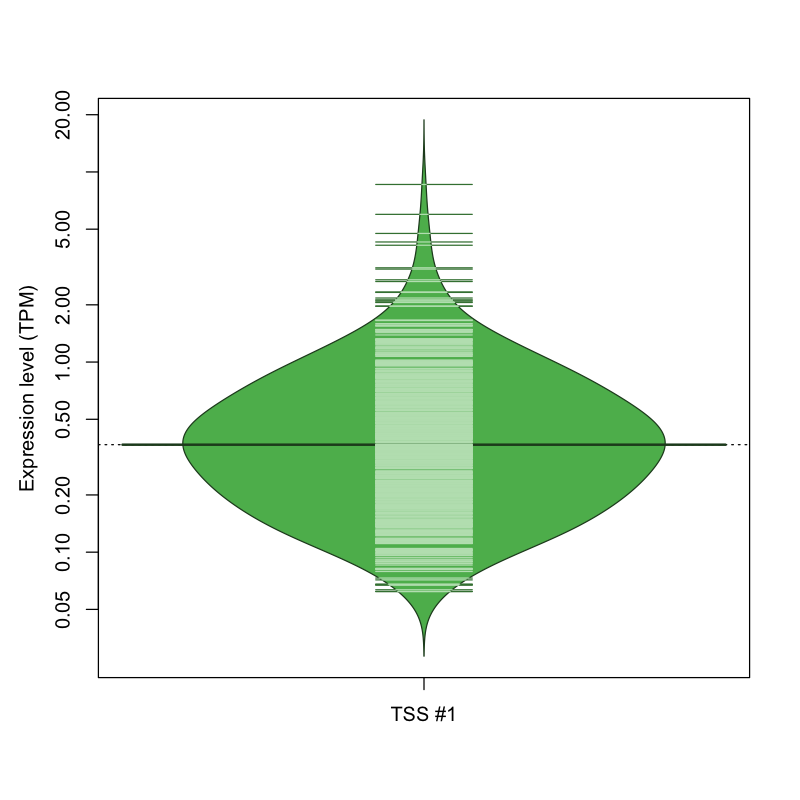
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_83981 | 1216 libraries | 546 libraries | 65 libraries | 2 libraries | 0 libraries |
The graphical summary, for retro_hsap_459 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_459 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_459 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 7 parental genes, and 7 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000010607 | 1 retrocopy | |
| Dasypus novemcinctus | ENSDNOG00000005103 | 1 retrocopy | |
| Equus caballus | ENSECAG00000022933 | 1 retrocopy | |
| Homo sapiens | ENSG00000118985 | 1 retrocopy |
retro_hsap_459 ,
|
| Nomascus leucogenys | ENSNLEG00000014192 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000015647 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000017090 | 1 retrocopy |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .06 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .13 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .07 RPM |
| CEU_NA12751 | 0 .05 RPM |
| CEU_NA12760 | 0 .09 RPM |
| CEU_NA12827 | 0 .05 RPM |
| CEU_NA12872 | 0 .08 RPM |
| CEU_NA12873 | 0 .06 RPM |
| FIN_HG00183 | 0 .08 RPM |
| FIN_HG00277 | 0 .04 RPM |
| FIN_HG00315 | 0 .06 RPM |
| FIN_HG00321 | 0 .06 RPM |
| FIN_HG00328 | 0 .02 RPM |
| FIN_HG00338 | 0 .02 RPM |
| FIN_HG00349 | 0 .06 RPM |
| FIN_HG00375 | 0 .07 RPM |
| FIN_HG00377 | 0 .10 RPM |
| FIN_HG00378 | 0 .04 RPM |
| GBR_HG00099 | 0 .12 RPM |
| GBR_HG00111 | 0 .04 RPM |
| GBR_HG00114 | 0 .05 RPM |
| GBR_HG00119 | 0 .05 RPM |
| GBR_HG00131 | 0 .09 RPM |
| GBR_HG00133 | 0 .05 RPM |
| GBR_HG00134 | 0 .04 RPM |
| GBR_HG00137 | 0 .05 RPM |
| GBR_HG00142 | 0 .03 RPM |
| GBR_HG00143 | 0 .03 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .11 RPM |
| TSI_NA20532 | 0 .07 RPM |
| TSI_NA20538 | 0 .14 RPM |
| TSI_NA20756 | 0 .09 RPM |
| TSI_NA20765 | 0 .12 RPM |
| TSI_NA20771 | 0 .06 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .03 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .03 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .03 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .03 RPM |
| YRI_NA19118 | 0 .04 RPM |
| YRI_NA19213 | 0 .10 RPM |
| YRI_NA19214 | 0 .02 RPM |
| YRI_NA19223 | 0 .02 RPM |