RetrogeneDB ID: | retro_mmus_11 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 1:16768269..16770138(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000091020 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tox4 | ||
| Ensembl ID: | ENSMUSG00000016831 | ||
| Aliases: | Tox4, 5730589K01Rik, A630040M18, AA410149, LCP1 | ||
| Description: | TOX high mobility group box family member 4 [Source:MGI Symbol;Acc:MGI:1915389] |

Retrocopy-Parental alignment summary:
>retro_mmus_11
ATGGAGTTTCCCAGAGGAAATGACAATTACCTGACAATCACTGGGCCCTCGCACCCTTTCCTGTCAGGGGCCGAGACATT
ACATACACCAAGCTTGGGCGATGAGGAATTTGAAATTCCACCTATCTCCTTGGACTCTGATCCCTCACTGGCTGTCTCAG
ATGTGGTTGGCCACTTTGATGACCTGGCAGACCACTCCTCTTCACAGGATAGCAGCTTTTCAGCTCAGTACGGGGTCCAG
ACATTGGACATGCCTGTGGGCATGACCCATGGCTTGATGGAGCAGGGCGGGGGGCTCCTGAGTGGGGGCTTGACTATGGA
CCTGGACCATTCTATAGGAACTCAGTATAATGCCAACCCACCTGCTACAATTGATGTACCAATGACAGACATGACATCTG
GCTTAATGGGACACAGCCAGTTGACCTCTATTGGTCAGTCAGAACTGAGTTCTCAACTTGGTTTGAGTTTAGGAGGTGGC
ACTATCTTGCCACCTGCCCAGTCACCTGAGGATCGTCTTTCAACTACTCCTTCACCTACCAGTTCACTTCATGAGGATGG
GGTTGATGATTTCCGGAGGCAACTTCCCGCCCAGAAGATAGTGGTAGTGGAAACAGGGAAAAAGCAAAAGGCTCCCAAGA
AGAGAAAAAAGAAAGACCCAAATGAACCGCAGAAGCCAGTCTCTGCTTATGCATTGCTCTTTCGAGATACACAGGCTGCC
ATCAAGGGACAAAACCCGAATGCAACCTTTGGGGAGGTATCGAAGATTGTGGCTTCCATGTGGGACAGTCTTGGAGAAGA
ACAGAAGCAGGTGTACAAGCGGAAGGCTGAGGCTGCCAAGAAAGAGTATCTCAAGGCACTAGCTGCTTATAAAGACAACC
AGGAATGCCAGGCTACTGTGGAAACCGTGGAATTAGATCCTGTGCCACAGTCACAGACTCCTTCCCCACCTCCTGTGACT
ACTGCTGACCCAGCATCTCCAGCACCAGCCTCAACGGAGTCTCCTGCTTTGCCCCCTTGCATCATAGTGAATTCCATGCT
TTCATCTAATGTGGCAAACCAAGCCTCTTCTGGGCCTGGAGGTCAGCCCAATATTACCAAGTTGATTATTACCAAACAGA
TGTTGCCTTCTTCTATCACCATGTCCCAAGGAGGAATGGTTACTGTTATCCCAGGCACAGTGGTTACCTCCCGTGGTCTC
CAGGTAGGCCAAACAAGTACAGCTACTATTCAGCCAAGCCAGCAGGCCCAGATTGTCACCCGGTCAGTGTTGCAGGCAGC
AGCAGCTGCTGCTGCTTCCATGCAACTGCCTCCACCCCGACTACAGCCCCCTCCGTTGCAGCCCCCTCCGTTGCAACAGA
TGCCTAAGCCCCCAATTCAACAGCAAATCACCATTCTACAGCAACCCCCTCCACTTCAGGCCATGCAGCAACCTCCACCT
CAGAAAGTTAGAATCAACTTACAGCAGCAGCCACCTCCTCTGCAGAGCAAGATTGTGCCTCCACCCACTCTGAAAATACA
GACTACTGTGGTCCTTCCAACTGTAGAAAGCAGTCCTGAGCAGCCTATGAACAGCAGCCCCGAGGCCCACACAGTAGAGG
CAACCTCTCCTGAGACAATCTGTGAAATGATCGAAGTAGTTCCTGAGGTGGAGTCTCCTTCTCAGATGAATGTTGAATTG
GTGAGTGGCTCCCCTGTGACACTGTCACCTCAGCCTGGATGTGTGAGGTCTAGTTGTGAGAACCCTCCTGTTATCAGTAA
GGACTGGGACAACGAGTACTGCAACAATGAGTGTGTGGTGAAGCACTGCAGGGATGTATTCTTGGCCTGGGTGGCCTCTA
GAAATCCAAACTCTGGTGTTTGTGAGGTA
ATGGAGTTTCCCAGAGGAAATGACAATTACCTGACAATCACTGGGCCCTCGCACCCTTTCCTGTCAGGGGCCGAGACATT
ACATACACCAAGCTTGGGCGATGAGGAATTTGAAATTCCACCTATCTCCTTGGACTCTGATCCCTCACTGGCTGTCTCAG
ATGTGGTTGGCCACTTTGATGACCTGGCAGACCACTCCTCTTCACAGGATAGCAGCTTTTCAGCTCAGTACGGGGTCCAG
ACATTGGACATGCCTGTGGGCATGACCCATGGCTTGATGGAGCAGGGCGGGGGGCTCCTGAGTGGGGGCTTGACTATGGA
CCTGGACCATTCTATAGGAACTCAGTATAATGCCAACCCACCTGCTACAATTGATGTACCAATGACAGACATGACATCTG
GCTTAATGGGACACAGCCAGTTGACCTCTATTGGTCAGTCAGAACTGAGTTCTCAACTTGGTTTGAGTTTAGGAGGTGGC
ACTATCTTGCCACCTGCCCAGTCACCTGAGGATCGTCTTTCAACTACTCCTTCACCTACCAGTTCACTTCATGAGGATGG
GGTTGATGATTTCCGGAGGCAACTTCCCGCCCAGAAGATAGTGGTAGTGGAAACAGGGAAAAAGCAAAAGGCTCCCAAGA
AGAGAAAAAAGAAAGACCCAAATGAACCGCAGAAGCCAGTCTCTGCTTATGCATTGCTCTTTCGAGATACACAGGCTGCC
ATCAAGGGACAAAACCCGAATGCAACCTTTGGGGAGGTATCGAAGATTGTGGCTTCCATGTGGGACAGTCTTGGAGAAGA
ACAGAAGCAGGTGTACAAGCGGAAGGCTGAGGCTGCCAAGAAAGAGTATCTCAAGGCACTAGCTGCTTATAAAGACAACC
AGGAATGCCAGGCTACTGTGGAAACCGTGGAATTAGATCCTGTGCCACAGTCACAGACTCCTTCCCCACCTCCTGTGACT
ACTGCTGACCCAGCATCTCCAGCACCAGCCTCAACGGAGTCTCCTGCTTTGCCCCCTTGCATCATAGTGAATTCCATGCT
TTCATCTAATGTGGCAAACCAAGCCTCTTCTGGGCCTGGAGGTCAGCCCAATATTACCAAGTTGATTATTACCAAACAGA
TGTTGCCTTCTTCTATCACCATGTCCCAAGGAGGAATGGTTACTGTTATCCCAGGCACAGTGGTTACCTCCCGTGGTCTC
CAGGTAGGCCAAACAAGTACAGCTACTATTCAGCCAAGCCAGCAGGCCCAGATTGTCACCCGGTCAGTGTTGCAGGCAGC
AGCAGCTGCTGCTGCTTCCATGCAACTGCCTCCACCCCGACTACAGCCCCCTCCGTTGCAGCCCCCTCCGTTGCAACAGA
TGCCTAAGCCCCCAATTCAACAGCAAATCACCATTCTACAGCAACCCCCTCCACTTCAGGCCATGCAGCAACCTCCACCT
CAGAAAGTTAGAATCAACTTACAGCAGCAGCCACCTCCTCTGCAGAGCAAGATTGTGCCTCCACCCACTCTGAAAATACA
GACTACTGTGGTCCTTCCAACTGTAGAAAGCAGTCCTGAGCAGCCTATGAACAGCAGCCCCGAGGCCCACACAGTAGAGG
CAACCTCTCCTGAGACAATCTGTGAAATGATCGAAGTAGTTCCTGAGGTGGAGTCTCCTTCTCAGATGAATGTTGAATTG
GTGAGTGGCTCCCCTGTGACACTGTCACCTCAGCCTGGATGTGTGAGGTCTAGTTGTGAGAACCCTCCTGTTATCAGTAA
GGACTGGGACAACGAGTACTGCAACAATGAGTGTGTGGTGAAGCACTGCAGGGATGTATTCTTGGCCTGGGTGGCCTCTA
GAAATCCAAACTCTGGTGTTTGTGAGGTA
ORF - retro_mmus_11 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.56 % |
| Parental protein coverage: | 99.52 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MEFPGGNDNYLTITGPSHPFLSGAETFHTPSLGDEEFEIPPISLDSDPSLAVSDVVGHFDDLADPSSSQD |
| MEFP.GNDNYLTITGPSHPFLSGAET.HTPSLGDEEFEIPPISLDSDPSLAVSDVVGHFDDLAD.SSSQD | |
| Retrocopy | MEFPRGNDNYLTITGPSHPFLSGAETLHTPSLGDEEFEIPPISLDSDPSLAVSDVVGHFDDLADHSSSQD |
| Parental | GSFSAQYGVQTLDMPVGMTHGLMEQGGGLLSGGLTMDLDHSIGTQYSANPPVTIDVPMTDMTSGLMGHSQ |
| .SFSAQYGVQTLDMPVGMTHGLMEQGGGLLSGGLTMDLDHSIGTQY.ANPP.TIDVPMTDMTSGLMGHSQ | |
| Retrocopy | SSFSAQYGVQTLDMPVGMTHGLMEQGGGLLSGGLTMDLDHSIGTQYNANPPATIDVPMTDMTSGLMGHSQ |
| Parental | LTTIDQSELSSQLGLSLGGGTILPPAQSPEDRLSTTPSPTNSLHEDGVDDFRRQLPAQKTVVVETGKKQK |
| LT.I.QSELSSQLGLSLGGGTILPPAQSPEDRLSTTPSPT.SLHEDGVDDFRRQLPAQK.VVVETGKKQK | |
| Retrocopy | LTSIGQSELSSQLGLSLGGGTILPPAQSPEDRLSTTPSPTSSLHEDGVDDFRRQLPAQKIVVVETGKKQK |
| Parental | APKKRKKKDPNEPQKPVSAYALFFRDTQAAIKGQNPNATFGEVSKIVASMWDSLGEEQKQVYKRKTEAAK |
| APKKRKKKDPNEPQKPVSAYAL.FRDTQAAIKGQNPNATFGEVSKIVASMWDSLGEEQKQVYKRK.EAAK | |
| Retrocopy | APKKRKKKDPNEPQKPVSAYALLFRDTQAAIKGQNPNATFGEVSKIVASMWDSLGEEQKQVYKRKAEAAK |
| Parental | KEYLKALAAYKDNQECQATVETVELDPVPQSQTPSPPPVTAADPASPAPASTESPALPPCIIVNSTLSSY |
| KEYLKALAAYKDNQECQATVETVELDPVPQSQTPSPPPVT.ADPASPAPASTESPALPPCIIVNS.LSS. | |
| Retrocopy | KEYLKALAAYKDNQECQATVETVELDPVPQSQTPSPPPVTTADPASPAPASTESPALPPCIIVNSMLSSN |
| Parental | VANQASSGPGGQPNITKLIITKQMLPSSITMSQGGMVTVIPATVVTSRGLQVGQTSTATIQPSQQAQIVT |
| VANQASSGPGGQPNITKLIITKQMLPSSITMSQGGMVTVIP.TVVTSRGLQVGQTSTATIQPSQQAQIVT | |
| Retrocopy | VANQASSGPGGQPNITKLIITKQMLPSSITMSQGGMVTVIPGTVVTSRGLQVGQTSTATIQPSQQAQIVT |
| Parental | RSVLQAAA-AAAASM----QLPPPRLQPPPLQQMPQPPTQQQVTILQQPPPLQAMQQPPPQKVRINLQQQ |
| RSVLQAAA.AAA........L.PP.LQPPPLQQMP.PP.QQQ.TILQQPPPLQAMQQPPPQKVRINLQQQ | |
| Retrocopy | RSVLQAAAAAAASMQLPPPRLQPPPLQPPPLQQMPKPPIQQQITILQQPPPLQAMQQPPPQKVRINLQQQ |
| Parental | PPPLQSKIVPPPTLKIQTTVVPPTVESSPEQPMNSSPEAHTVEATSPETICEMIADVVPEVESPSQMDVE |
| PPPLQSKIVPPPTLKIQTTVV.PTVESSPEQPMNSSPEAHTVEATSPETICEMI..VVPEVESPSQM.VE | |
| Retrocopy | PPPLQSKIVPPPTLKIQTTVVLPTVESSPEQPMNSSPEAHTVEATSPETICEMI-EVVPEVESPSQMNVE |
| Parental | LVSGSPVALSPQPRCVRSGCENPPVVSKDWDNEYCSNECVVKHCRDVFLAWVASRNPNSVV |
| LVSGSPV.LSPQP.CVRS.CENPPV.SKDWDNEYC.NECVVKHCRDVFLAWVASRNPNS.V | |
| Retrocopy | LVSGSPVTLSPQPGCVRSSCENPPVISKDWDNEYCNNECVVKHCRDVFLAWVASRNPNSGV |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .09 RPM | 60 .21 RPM |
| SRP007412_cerebellum | 1 .65 RPM | 62 .53 RPM |
| SRP007412_heart | 0 .09 RPM | 45 .01 RPM |
| SRP007412_kidney | 1 .33 RPM | 50 .20 RPM |
| SRP007412_liver | 0 .33 RPM | 32 .63 RPM |
| SRP007412_testis | 3 .16 RPM | 65 .04 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_11 was not detected
No EST(s) were mapped for retro_mmus_11 retrocopy.
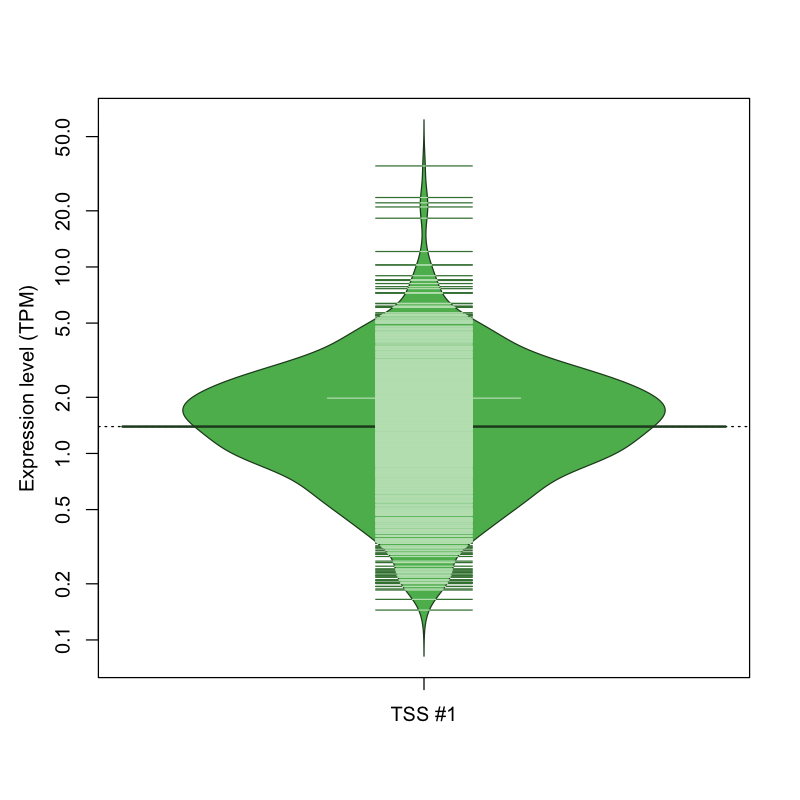
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_71379 | 164 libraries | 290 libraries | 581 libraries | 29 libraries | 8 libraries |
The graphical summary, for retro_mmus_11 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_11 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_11 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 11 parental genes, and 13 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000006261 | 1 retrocopy | |
| Canis familiaris | ENSCAFG00000005647 | 2 retrocopies | |
| Choloepus hoffmanni | ENSCHOG00000000714 | 2 retrocopies | |
| Homo sapiens | ENSG00000092203 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000010501 | 1 retrocopy | |
| Mustela putorius furo | ENSMPUG00000000181 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000016831 | 1 retrocopy |
retro_mmus_11 ,
|
| Nomascus leucogenys | ENSNLEG00000014556 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000005598 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000006126 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000029203 | 1 retrocopy |