RetrogeneDB ID: | retro_mmus_1130 | ||
Retrocopy location | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 13:89577157..89578413(-) | ||
| Located in intron of: | ENSMUSG00000021613 | ||
Retrocopy information | Ensembl ID: | ENSMUSG00000094463 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | Agk | ||
| Ensembl ID: | ENSMUSG00000029916 | ||
| Aliases: | Agk, 2610037M15Rik, 6720408I04Rik, AI465370, Mulk | ||
| Description: | acylglycerol kinase [Source:MGI Symbol;Acc:MGI:1917173] |

Retrocopy-Parental alignment summary:
>retro_mmus_1130
TTCTTTAAAACGCTTCGAAATCATTGGAAGAAAACTATAGCTGGGCTCTGTTTGCTGACATGGGGAGGTCACTGGCTCTA
CGGGAAGCACTGTGATAACCTGCTAAGAAGAGCAGCCTGTCAAGAAGCTCAGGTGTTTGGCAACCAGCTCATTCCTCCCA
ATGCACAAGTGAAGAAAGCCACCGTCTTTCTCAATCCCGCAGCTTGCAAAGGCAAAGCCAGAACTCTATTTGAAAAAAAT
ACTGCTCCAATTTTACACCTGTCTGGCATGGATGTGACTGTTGTCAAGATGGATTATGAGGGACAAGCCAAGAAGCTCCT
GGAGTTGATGGAAAGCACTGATGTGATCATTGTTGCTGGAGGAGACGGCACACTGCAGGAGGTTGTTACTGGAGTACTGA
GAAGAACAGATGAGGCTACCTTCAGTAAGATTCCAATTGGATTTATCCCGCTGGGACAGACCAGTAGTTTGAGTCACACT
CTCTTTGCTGAAAGTGGAAACAAAGTCCAACATATTTTCAGACGCCACACTTGCCATTGTGAAAGGAGAGACTGTTCCAC
TTGATGTCTTGCAGATCAAGGGTGAAAAAGAACAACCTGTGTATGCAATGACTGGCCTCCGGTGGGGGATCCTTCAGAGA
TGCTGGCGTCAAAGTTAGCAAGTACTGGTATCTTGGTCCACTAAAAACCAAAGCAGCCCACTTTTTCAGCACTCTTCAGG
AATGGCCTCAGACTCATCAAGCCTCCATCTCTTACACGGGACCTAGAGAGAGACCTCCCATTGAGCCTGAAGAGACTCCT
CCCAGGCCGTCTCTGTATAGGAGAATATTACGCAGGCTTGCCTCATTCTGGGCACAGCCACAGGATGCCTCCTCCCGAGA
GGTAAGCCCTGAGGTCTGGAAAGATGTACAACTGTCCACCATTGAACTGTCCATCACAACCCGAAACACCCAGCTTGACC
TGATGAGCAAAGAGGATTTTATGAACATTTGCATTGAACCTGACACTGTCAGCAAAGGAGATTTCATAATCATAGGAAGT
AAGAAGGTGAGAGACCCTGGCCTGCGTGCAGCTGGCACCGAGTGCCTCCAAGCCAGCCACTGTACTCTGGTTCTTCCTGA
GGGAACTGAGGGCTCCTTCAGCATTGACAGCGAGGAGTACGAAGCCATGCCGGTGGAGGTCAAACTACTGCCCAGAAAGC
TGCGGTTCTTCTGTGATCCGAGGAAGAGAGAGCAAATGCTGCCGAGCACATCCCAG
ORF - retro_mmus_1130 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 98.57 % |
| Parental protein coverage: | 99.29 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | FFKTLRNHWKKTTAGLCLLTWGGHWLYGKHCDNLLRRAACQEAQVFGNQLIPPNAQVKKATVFLNPAACK |
| FFKTLRNHWKKT.AGLCLLTWGGHWLYGKHCDNLLRRAACQEAQVFGNQLIPPNAQVKKATVFLNPAACK | |
| Retrocopy | FFKTLRNHWKKTIAGLCLLTWGGHWLYGKHCDNLLRRAACQEAQVFGNQLIPPNAQVKKATVFLNPAACK |
| Parental | GKARTLFEKNAAPILHLSGMDVTVVKTDYEGQAKKLLELMESTDVIIVAGGDGTLQEVVTGVLRRTDEAT |
| GKARTLFEKN.APILHLSGMDVTVVK.DYEGQAKKLLELMESTDVIIVAGGDGTLQEVVTGVLRRTDEAT | |
| Retrocopy | GKARTLFEKNTAPILHLSGMDVTVVKMDYEGQAKKLLELMESTDVIIVAGGDGTLQEVVTGVLRRTDEAT |
| Parental | FSKIPIGFIPLGQTSSLSHTLFAESGNKVQHI-TDATLAIVKGETVPLDVLQIKGEKEQPVYAMTGLRW- |
| FSKIPIGFIPLGQTSSLSHTLFAESGNKVQHI..DATLAIVKGETVPLDVLQIKGEKEQPVYAMTGLRW. | |
| Retrocopy | FSKIPIGFIPLGQTSSLSHTLFAESGNKVQHI>SDATLAIVKGETVPLDVLQIKGEKEQPVYAMTGLRW> |
| Parental | GSFRDAGVKVSKYWYLGPLKTKAAHFFSTLQEWPQTHQASISYTGPRERPPIEPEETPPRPSLYRRILRR |
| GSFRDAGVKVSKYWYLGPLKTKAAHFFSTLQEWPQTHQASISYTGPRERPPIEPEETPPRPSLYRRILRR | |
| Retrocopy | GSFRDAGVKVSKYWYLGPLKTKAAHFFSTLQEWPQTHQASISYTGPRERPPIEPEETPPRPSLYRRILRR |
| Parental | LASFWAQPQDASSREVSPEVWKDVQLSTIELSITTRNTQLDLMSKEDFMNICIEPDTVSKGDFIIIGSKK |
| LASFWAQPQDASSREVSPEVWKDVQLSTIELSITTRNTQLDLMSKEDFMNICIEPDTVSKGDFIIIGSKK | |
| Retrocopy | LASFWAQPQDASSREVSPEVWKDVQLSTIELSITTRNTQLDLMSKEDFMNICIEPDTVSKGDFIIIGSKK |
| Parental | VRDPGLRAAGTECLQASHCTLVLPEGTEGSFSIDSEEYEAMPVEVKLLPRKLRFFCDPRKREQMLPSTSQ |
| VRDPGLRAAGTECLQASHCTLVLPEGTEGSFSIDSEEYEAMPVEVKLLPRKLRFFCDPRKREQMLPSTSQ | |
| Retrocopy | VRDPGLRAAGTECLQASHCTLVLPEGTEGSFSIDSEEYEAMPVEVKLLPRKLRFFCDPRKREQMLPSTSQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .61 RPM | 12 .55 RPM |
| SRP007412_cerebellum | 1 .13 RPM | 9 .94 RPM |
| SRP007412_heart | 0 .19 RPM | 7 .81 RPM |
| SRP007412_kidney | 0 .58 RPM | 5 .99 RPM |
| SRP007412_liver | 0 .22 RPM | 2 .40 RPM |
| SRP007412_testis | 0 .50 RPM | 3 .52 RPM |
RNA Polymerase II activity near the 5' end of retro_mmus_1130 was not detected
No EST(s) were mapped for retro_mmus_1130 retrocopy.
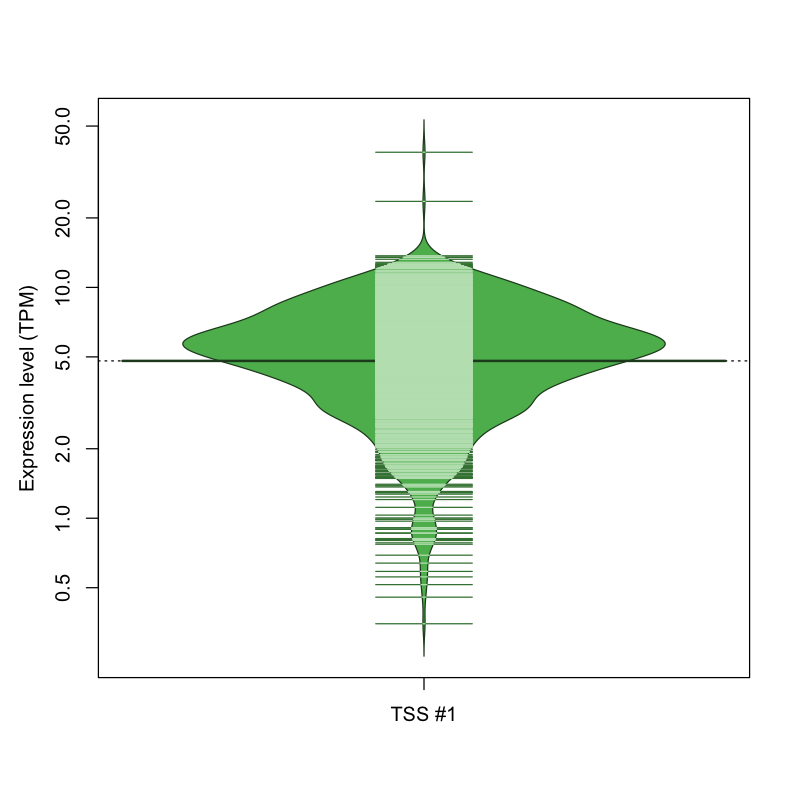
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_31434 | 47 libraries | 22 libraries | 441 libraries | 492 libraries | 70 libraries |
The graphical summary, for retro_mmus_1130 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_1130 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_1130 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 9 parental genes, and 10 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000013589 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000008821 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000002479 | 1 retrocopy | |
| Microcebus murinus | ENSMICG00000002561 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000021017 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000029916 | 1 retrocopy |
retro_mmus_1130 ,
|
| Nomascus leucogenys | ENSNLEG00000002291 | 1 retrocopy | |
| Pongo abelii | ENSPPYG00000018064 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000019775 | 2 retrocopies |