RetrogeneDB ID: | retro_mmus_1433 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 16:8612970..8613408(+) | ||
| Located in intron of: | ENSMUSG00000057880 | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Zmynd19 | ||
| Ensembl ID: | ENSMUSG00000026974 | ||
| Aliases: | None | ||
| Description: | zinc finger, MYND domain containing 19 [Source:MGI Symbol;Acc:MGI:1914437] |

Retrocopy-Parental alignment summary:
>retro_mmus_1433
AAAAACAAACAGGACTTGAGTGGGCTTCTCTATGAGATCCTGTGGGAACCACACAGGGAGGCATGGCCCACTGGTTTCAG
GTAGAGCATCTCAATATTGTGACAGAGGATGTTTGCCTGGAAAACCTGCAGTTGATGCTGTGGAACCGGCAGACCAAAGC
TAAAGAGACCTCCAGTAACAAGGGAGCAAAGCTTATACTGGCTGGCAACCCAGCAGCTTTCCACGGACCCCAGAGAAGAG
CAGTCCCCTCTCCTGGATGTGACTCAGCACCGCAGTGCCAGTGGGGTGTGGTGGAGGAGAATTCCTGGAGCCATTCTGAG
TGTCATCATGTTCCCTGCAGTGAGACTGAGAAGCAGCTTCCCAAGATCAGCCTTTGTGGATGCTGCCAGGTGGCCTGGGA
CCATGGGTCCCAGGTGCCAGCAGCAGGATTGGATTGCC
AAAAACAAACAGGACTTGAGTGGGCTTCTCTATGAGATCCTGTGGGAACCACACAGGGAGGCATGGCCCACTGGTTTCAG
GTAGAGCATCTCAATATTGTGACAGAGGATGTTTGCCTGGAAAACCTGCAGTTGATGCTGTGGAACCGGCAGACCAAAGC
TAAAGAGACCTCCAGTAACAAGGGAGCAAAGCTTATACTGGCTGGCAACCCAGCAGCTTTCCACGGACCCCAGAGAAGAG
CAGTCCCCTCTCCTGGATGTGACTCAGCACCGCAGTGCCAGTGGGGTGTGGTGGAGGAGAATTCCTGGAGCCATTCTGAG
TGTCATCATGTTCCCTGCAGTGAGACTGAGAAGCAGCTTCCCAAGATCAGCCTTTGTGGATGCTGCCAGGTGGCCTGGGA
CCATGGGTCCCAGGTGCCAGCAGCAGGATTGGATTGCC
ORF - retro_mmus_1433 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 56.49 % |
| Parental protein coverage: | 66.08 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 4 |
Retrocopy - Parental Gene Alignment:
| Parental | KNRGRGSGRLLHELLWERHR-GGVAPGFQVVHLNAVTVDNRLDNLQLVPWG-WRPKAEETSSKQREQSLY |
| KN....SG.LL.E.LWE.HR.GG.A..FQV.HLN.VT.D..L.NLQL..W...RPK........REQSLY | |
| Retrocopy | KNKQDLSG-LLYEILWEPHR<GGMAHWFQVEHLNIVTEDVCLENLQLMLWN>GRPKLKRPPVT-REQSLY |
| Parental | WLAIQQLPTDPIEEQFPVLNVTRYYNANG-DVVEEEENSCTYYECHYPPCTVIEKQLREFNICGRCQVAR |
| WLA.QQL.TDP.EEQ.P.L.VT....A.G..VVE..ENS....ECH..PC...EKQL.....CG.CQVA. | |
| Retrocopy | WLATQQLSTDPREEQSPLLDVTQHRSASG<GVVE--ENSWSHSECHHVPCSETEKQLPKISLCGCCQVAW |
| Parental | YCGSQ-CQQKDWPA |
| ..GSQ.CQQ.DW.A | |
| Retrocopy | DHGSQ>CQQQDWIA |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .86 RPM | 21 .56 RPM |
| SRP007412_cerebellum | 0 .43 RPM | 20 .10 RPM |
| SRP007412_heart | 0 .00 RPM | 12 .79 RPM |
| SRP007412_kidney | 0 .21 RPM | 10 .30 RPM |
| SRP007412_liver | 0 .16 RPM | 11 .41 RPM |
| SRP007412_testis | 0 .14 RPM | 44 .37 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_1433 was not detected
No EST(s) were mapped for retro_mmus_1433 retrocopy.
No TSS is located nearby retro_mmus_1433 retrocopy 5' end.
| Experiment type: | PCR amplification |
|---|---|
| Forward primer: | GCTAGAGGAGAGATACTCGT (20 nt long) |
| Reverse primer: | GAGCACTGGAGATAGAGTCC (20 nt long) |
| Anneling temperature: | 55 °C |
| (Expected) product size: | 732 |
| Electrophoresis gel image: | 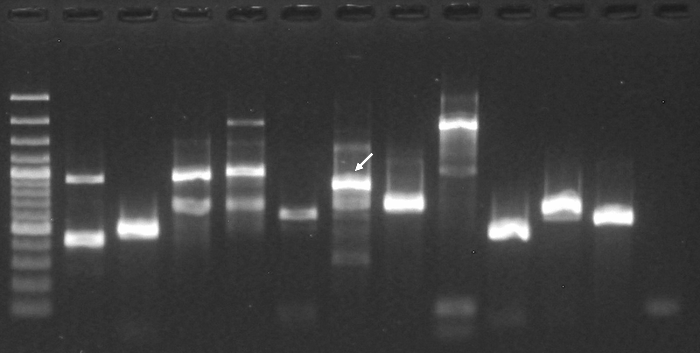 |
| Additional comment: | Pfu DNA Polymerase; pooled mouse cDNA; 40 cycles of PCR |
Retrocopy orthology:
Retrocopy retro_mmus_1433 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 12 parental genes, and 15 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Dasypus novemcinctus | ENSDNOG00000006644 | 2 retrocopies | |
| Dipodomys ordii | ENSDORG00000008360 | 1 retrocopy | |
| Equus caballus | ENSECAG00000012412 | 1 retrocopy | |
| Echinops telfairi | ENSETEG00000005023 | 1 retrocopy | |
| Homo sapiens | ENSG00000165724 | 1 retrocopy | |
| Gorilla gorilla | ENSGGOG00000006875 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000011727 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000019610 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000017172 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000026974 | 3 retrocopies |
retro_mmus_1433 , retro_mmus_2466, retro_mmus_942,
|
| Nomascus leucogenys | ENSNLEG00000009236 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000010812 | 1 retrocopy |