RetrogeneDB ID: | retro_mmus_535 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 10:22343438..22345478(+) | ||
| Located in intron of: | ENSMUSG00000053219 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000097245 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Aco2 | ||
| Ensembl ID: | ENSMUSG00000022477 | ||
| Aliases: | Aco2, Aco-2, Aco3, D10Wsu183e | ||
| Description: | aconitase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:87880] |

Retrocopy-Parental alignment summary:
>retro_mmus_535
ATGGCTCCTTACAGCCTCCTGGTCACCCGGCTGCAGAAGGCCCTGGGTGTGGGGCAGTACCATGTGGCCTCTGTCCTGTG
CCAAAGGGCCAAGGTGGCCATGAGCCATTTTGAGCCCAGTGAGTACATCCGATATGACCTGCTAGAGAAGAACATTAACA
TTGTCTGTAAACGGTTGAACCGGCCTCTTACTCTCTCAGAGAAGATTGTGTATGGACACCTGGATGACCCAGCCAATTGG
GAGATCGAGCAGGGAAAGACATATCTGCGTTTATGGCCTGACCGGGTGGCCATGCAGGATGCCACAGGACAGATGGCTAT
GCTACAGTTCATTAGCAGTGGGCTGCCCAAGGTGGCTGTACCATCAACCATCCACTGTGATCATCTGATTGAGGCCCAGG
TTGGGGGTGAGAAAGACCTGCGCAGGGCCAAGGACATAAACCAGGAAGTGTATAATTTCCTGGCAACTGCAGGTGCCAAG
TATGGCGTGGGCTTCTGGACGCCTGGAACTGGGATCATTTACCAGATCATTCTAGAAAACTATGCATACCCTGGAGTTCT
TCTGATTGGCACTGACTCGCACACCCCCAATGGTGGTGGCCTGGGAGGCATCTGCATTAGAGTAAGGGGTGCTGATGCTG
TGGATGTTATGGCTGGGATCCCCTGGGAGCTGAAGTGTCCCAAGGTGATTGGTGTGAAGCTGACAGGCTCCCTCTCTAGT
TGGACCTCACCCAAAGATGTGATCCTGAAACTGGCAGGTATCCTCACAGTGAAAGGTGGCACAGGTGCTATTGTGGAATA
CCATGGACCCGGTGTCGATTCCATCTCCTGCACTGGCATGGCAACTATCTGCAACATGGGTGCAGAAATTGGGGCCACTA
TATCAGTGTTCCCATACAACCACAGGATGAAAGGTACCTGAGCAAGACAGGCCGAACAGACATTGCCAACCTAGCAGAAG
AATTCAAGGATCACTTAGTGCCTGATCCTGGCTGCCAGTATGACCAAGTGATTGAAATTAACTTCAATGAGCTAAAGCCA
CATATCAATGGGCCCTTTACCCCTGACTTGGCTCATCCAGTGGCAGATGTGGGGACTGTGGCAGAGAAGGAAGGCTGGCC
TCTGGACATCTGAGTAGGTTTGATTGGCAGCTGCACCAATTCAATCTACGAGGACATGGGACGATCAGCAGCTGTGGCTA
AGCAGGCATTGGTCCATGGACTCAAGTGCAAGCCCCAGTTCACCATCACCCCAGGCTCTGAGCAGATCCGTGCTGCTATT
GAGCGGGATGGCTATGCACAGATCCTAAGGGATGTGGGTGGAATCGTTTTGGCCAATGCCTGTGGCTCTTGCATTGGCCA
GTGGGACAGAAAAGACATCAAGAGGGGGGAGAAGAATACAATCGTCACCTTCTACAACAAGAACTTCATAGGCCGTAATG
ATGCAAACCCTGAGACCCATGCCTTTGTCACATCCCCAGAGATTGTCACAGCTCTGGCCATTGCGGGGAAGCCTGAAGTT
CAATCCAGAAACTGACTTCTTGACAGGCAAGGATGGCAAGAAGTTCAAGTTGGAGGCTCCAGATGCAGACGAGCTTCCCC
GATCGGACTTTGATCCAGGGCAGGACACATACCAGCACCCCCCTAAGGACAGCAGTGGGCAGCGGGTGGATGTGAGCCCT
ACCAGCCAGCACCTACAGCTCCTGGAGCCTTTTGACAAGTGGGACGGCAAAGACCTTGAGGACCTGCAAATCCTCATCAA
GGTCAAAGGGAAGTGCACCACAGACCACATTTCTGCTGCCAGCCCCTGGCTCAAGTTCCGTGGTCATCTGGATAACATCT
CTAACAACCTGCTCATCGGTGCCATCAACATCGAAAATGGTAAAGCCAACTCTGTACGAAATGCTGTCACCCAGGAGTTT
GGCCCTGTACCTGACACTGCTCGCTACTACAAGAAACATGGCATCAGGTGGGTGGTGATTGGAGATGAGAACGATGGTGA
GGGCTCAAGCTGGGAGCATGCAGCACTGGAGCCTCGCCAT
ATGGCTCCTTACAGCCTCCTGGTCACCCGGCTGCAGAAGGCCCTGGGTGTGGGGCAGTACCATGTGGCCTCTGTCCTGTG
CCAAAGGGCCAAGGTGGCCATGAGCCATTTTGAGCCCAGTGAGTACATCCGATATGACCTGCTAGAGAAGAACATTAACA
TTGTCTGTAAACGGTTGAACCGGCCTCTTACTCTCTCAGAGAAGATTGTGTATGGACACCTGGATGACCCAGCCAATTGG
GAGATCGAGCAGGGAAAGACATATCTGCGTTTATGGCCTGACCGGGTGGCCATGCAGGATGCCACAGGACAGATGGCTAT
GCTACAGTTCATTAGCAGTGGGCTGCCCAAGGTGGCTGTACCATCAACCATCCACTGTGATCATCTGATTGAGGCCCAGG
TTGGGGGTGAGAAAGACCTGCGCAGGGCCAAGGACATAAACCAGGAAGTGTATAATTTCCTGGCAACTGCAGGTGCCAAG
TATGGCGTGGGCTTCTGGACGCCTGGAACTGGGATCATTTACCAGATCATTCTAGAAAACTATGCATACCCTGGAGTTCT
TCTGATTGGCACTGACTCGCACACCCCCAATGGTGGTGGCCTGGGAGGCATCTGCATTAGAGTAAGGGGTGCTGATGCTG
TGGATGTTATGGCTGGGATCCCCTGGGAGCTGAAGTGTCCCAAGGTGATTGGTGTGAAGCTGACAGGCTCCCTCTCTAGT
TGGACCTCACCCAAAGATGTGATCCTGAAACTGGCAGGTATCCTCACAGTGAAAGGTGGCACAGGTGCTATTGTGGAATA
CCATGGACCCGGTGTCGATTCCATCTCCTGCACTGGCATGGCAACTATCTGCAACATGGGTGCAGAAATTGGGGCCACTA
TATCAGTGTTCCCATACAACCACAGGATGAAAGGTACCTGAGCAAGACAGGCCGAACAGACATTGCCAACCTAGCAGAAG
AATTCAAGGATCACTTAGTGCCTGATCCTGGCTGCCAGTATGACCAAGTGATTGAAATTAACTTCAATGAGCTAAAGCCA
CATATCAATGGGCCCTTTACCCCTGACTTGGCTCATCCAGTGGCAGATGTGGGGACTGTGGCAGAGAAGGAAGGCTGGCC
TCTGGACATCTGAGTAGGTTTGATTGGCAGCTGCACCAATTCAATCTACGAGGACATGGGACGATCAGCAGCTGTGGCTA
AGCAGGCATTGGTCCATGGACTCAAGTGCAAGCCCCAGTTCACCATCACCCCAGGCTCTGAGCAGATCCGTGCTGCTATT
GAGCGGGATGGCTATGCACAGATCCTAAGGGATGTGGGTGGAATCGTTTTGGCCAATGCCTGTGGCTCTTGCATTGGCCA
GTGGGACAGAAAAGACATCAAGAGGGGGGAGAAGAATACAATCGTCACCTTCTACAACAAGAACTTCATAGGCCGTAATG
ATGCAAACCCTGAGACCCATGCCTTTGTCACATCCCCAGAGATTGTCACAGCTCTGGCCATTGCGGGGAAGCCTGAAGTT
CAATCCAGAAACTGACTTCTTGACAGGCAAGGATGGCAAGAAGTTCAAGTTGGAGGCTCCAGATGCAGACGAGCTTCCCC
GATCGGACTTTGATCCAGGGCAGGACACATACCAGCACCCCCCTAAGGACAGCAGTGGGCAGCGGGTGGATGTGAGCCCT
ACCAGCCAGCACCTACAGCTCCTGGAGCCTTTTGACAAGTGGGACGGCAAAGACCTTGAGGACCTGCAAATCCTCATCAA
GGTCAAAGGGAAGTGCACCACAGACCACATTTCTGCTGCCAGCCCCTGGCTCAAGTTCCGTGGTCATCTGGATAACATCT
CTAACAACCTGCTCATCGGTGCCATCAACATCGAAAATGGTAAAGCCAACTCTGTACGAAATGCTGTCACCCAGGAGTTT
GGCCCTGTACCTGACACTGCTCGCTACTACAAGAAACATGGCATCAGGTGGGTGGTGATTGGAGATGAGAACGATGGTGA
GGGCTCAAGCTGGGAGCATGCAGCACTGGAGCCTCGCCAT
ORF - retro_mmus_535 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 95.16 % |
| Parental protein coverage: | 87.18 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 2 |
Retrocopy - Parental Gene Alignment:
| Parental | MAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPSEYIRYDLLEKNINIVRKRLNRPLTLSEKIV |
| MAPYSLLVTRLQKALGV.QYHVASVLCQRAKVAMSHFEPSEYIRYDLLEKNINIV.KRLNRPLTLSEKIV | |
| Retrocopy | MAPYSLLVTRLQKALGVGQYHVASVLCQRAKVAMSHFEPSEYIRYDLLEKNINIVCKRLNRPLTLSEKIV |
| Parental | YGHLDDPANQEIERGKTYLRLRPDRVAMQDATAQMAMLQFISSGLPKVAVPSTIHCDHLIEAQVGGEKDL |
| YGHLDDPAN.EIE.GKTYLRL.PDRVAMQDAT.QMAMLQFISSGLPKVAVPSTIHCDHLIEAQVGGEKDL | |
| Retrocopy | YGHLDDPANWEIEQGKTYLRLWPDRVAMQDATGQMAMLQFISSGLPKVAVPSTIHCDHLIEAQVGGEKDL |
| Parental | RRAKDINQEVYNFLATAGAKYGVGFWRPGSGIIHQIILENYAYPGVLLIGTDSHTPNGGGLGGICIGVGG |
| RRAKDINQEVYNFLATAGAKYGVGFW.PG.GII.QIILENYAYPGVLLIGTDSHTPNGGGLGGICI.V.G | |
| Retrocopy | RRAKDINQEVYNFLATAGAKYGVGFWTPGTGIIYQIILENYAYPGVLLIGTDSHTPNGGGLGGICIRVRG |
| Parental | ADAVDVMAGIPWELKCPKVIGVKLTGSLSGWTSPKDVILKVAGILTVKGGTGAIVEYHGPGVDSISCTGM |
| ADAVDVMAGIPWELKCPKVIGVKLTGSLS.WTSPKDVILK.AGILTVKGGTGAIVEYHGPGVDSISCTGM | |
| Retrocopy | ADAVDVMAGIPWELKCPKVIGVKLTGSLSSWTSPKDVILKLAGILTVKGGTGAIVEYHGPGVDSISCTGM |
| Parental | ATICNMGAEIGATTSVFPYNHRMK-KYLSKTGRTDIANLAEEFKDHLVPDPGCQYDQVIEINLNELKPHI |
| ATICNMGAEIGAT.SVFPYNHRMK..YLSKTGRTDIANLAEEFKDHLVPDPGCQYDQVIEIN.NELKPHI | |
| Retrocopy | ATICNMGAEIGATISVFPYNHRMK<RYLSKTGRTDIANLAEEFKDHLVPDPGCQYDQVIEINFNELKPHI |
| Parental | NGPFTPDLAHPVADVGTVAEKEGWPLDIRVGLIGSCTNSSYEDMGRSAAVAKQALAHGLKCKSQFTITPG |
| NGPFTPDLAHPVADVGTVAEKEGWPLDI.VGLIGSCTNS.YEDMGRSAAVAKQAL.HGLKCK.QFTITPG | |
| Retrocopy | NGPFTPDLAHPVADVGTVAEKEGWPLDI*VGLIGSCTNSIYEDMGRSAAVAKQALVHGLKCKPQFTITPG |
| Parental | SEQIRATIERDGYAQILRDVGGIVLANACGPCIGQWDRKDIKKGEKNTIVTSYNRNFTGRNDANPETHAF |
| SEQIRA.IERDGYAQILRDVGGIVLANACG.CIGQWDRKDIK.GEKNTIVT.YN.NF.GRNDANPETHAF | |
| Retrocopy | SEQIRAAIERDGYAQILRDVGGIVLANACGSCIGQWDRKDIKRGEKNTIVTFYNKNFIGRNDANPETHAF |
| Parental | VTSPEIVTALAIA-GTLKFNPETDFLTGKDGKKFKLEAPDADELPRSDFDPGQDTYQHPPKDSSGQRVDV |
| VTSPEIVTALAIA.G.LKFNPETDFLTGKDGKKFKLEAPDADELPRSDFDPGQDTYQHPPKDSSGQRVDV | |
| Retrocopy | VTSPEIVTALAIA>GSLKFNPETDFLTGKDGKKFKLEAPDADELPRSDFDPGQDTYQHPPKDSSGQRVDV |
| Parental | SPTSQRLQLLEPFDKWDGKDLEDLQILIKVKGKCTTDHISAAGPWLKFRGHLDNISNNLLIGAINIENGK |
| SPTSQ.LQLLEPFDKWDGKDLEDLQILIKVKGKCTTDHISAA.PWLKFRGHLDNISNNLLIGAINIENGK | |
| Retrocopy | SPTSQHLQLLEPFDKWDGKDLEDLQILIKVKGKCTTDHISAASPWLKFRGHLDNISNNLLIGAINIENGK |
| Parental | ANSVRNAVTQEFGPVPDTARYYKKHGIRWVVIGDENYGEGSSREHAALEPRH |
| ANSVRNAVTQEFGPVPDTARYYKKHGIRWVVIGDEN.GEGSS.EHAALEPRH | |
| Retrocopy | ANSVRNAVTQEFGPVPDTARYYKKHGIRWVVIGDENDGEGSSWEHAALEPRH |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .68 RPM | 403 .43 RPM |
| SRP007412_cerebellum | 0 .87 RPM | 495 .96 RPM |
| SRP007412_heart | 4 .58 RPM | 2501 .31 RPM |
| SRP007412_kidney | 2 .71 RPM | 1359 .28 RPM |
| SRP007412_liver | 0 .41 RPM | 243 .59 RPM |
| SRP007412_testis | 0 .18 RPM | 147 .33 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_535 was not detected
No EST(s) were mapped for retro_mmus_535 retrocopy.
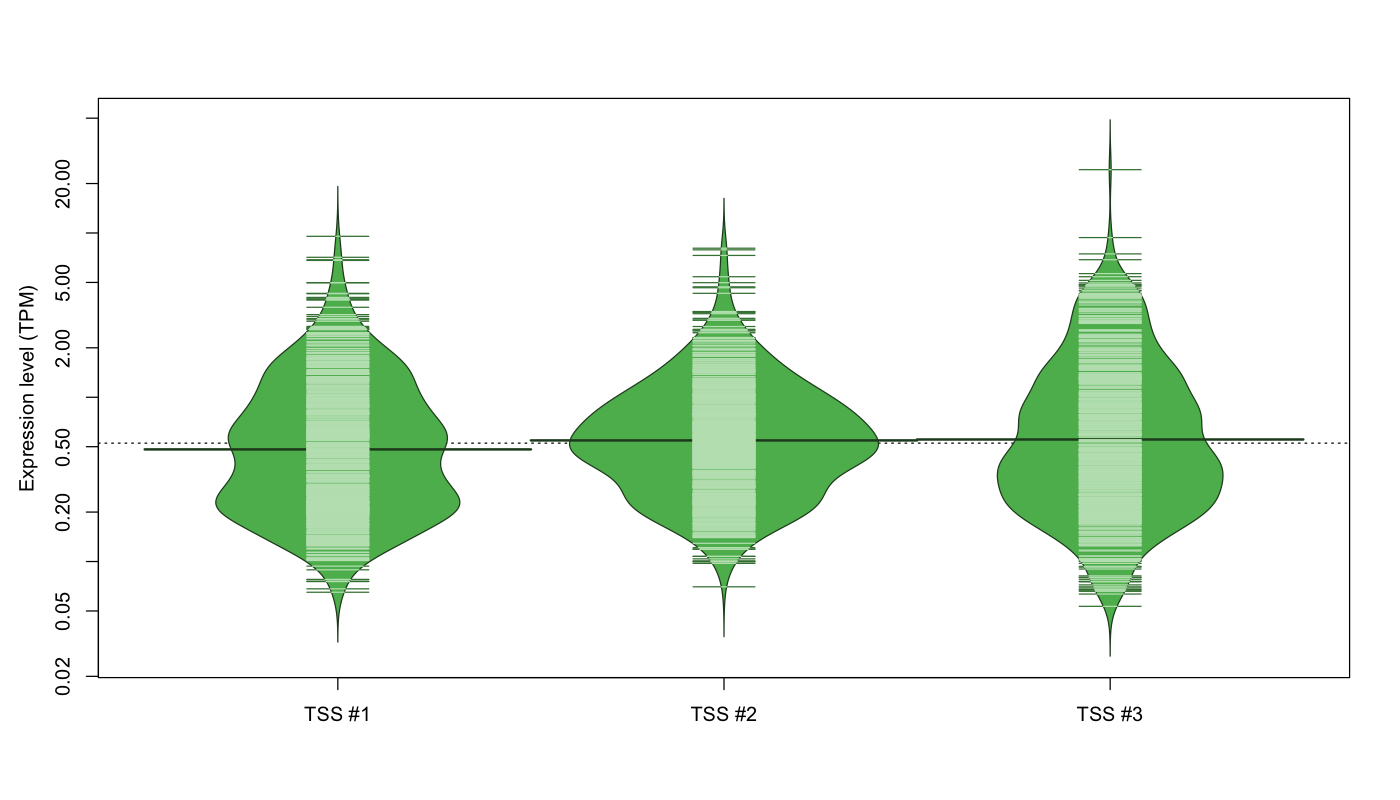
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_2495 | 468 libraries | 467 libraries | 133 libraries | 4 libraries | 0 libraries |
| TSS #2 | TSS_2496 | 423 libraries | 509 libraries | 136 libraries | 4 libraries | 0 libraries |
| TSS #3 | TSS_2497 | 516 libraries | 400 libraries | 149 libraries | 6 libraries | 1 library |
The graphical summary, for retro_mmus_535 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_535 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_535 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 8 parental genes, and 11 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Homo sapiens | ENSG00000100412 | 2 retrocopies | |
| Myotis lucifugus | ENSMLUG00000002472 | 1 retrocopy | |
| Macaca mulatta | ENSMMUG00000001454 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000022477 | 1 retrocopy |
retro_mmus_535 ,
|
| Nomascus leucogenys | ENSNLEG00000015482 | 2 retrocopies | |
| Pongo abelii | ENSPPYG00000011871 | 1 retrocopy | |
| Pan troglodytes | ENSPTRG00000014428 | 1 retrocopy | |
| Drosophila melanogaster | FBgn0010100 | 1 retrocopy |