RetrogeneDB ID: | retro_mmus_621 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 10:16424169..16424668(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | None | |
| Aliases: | None | ||
| Status: | NOVEL | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Sgta | ||
| Ensembl ID: | ENSMUSG00000004937 | ||
| Aliases: | Sgta, 5330427H01Rik, AI194281, D10Ertd190e, Sgt, Stg | ||
| Description: | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha [Source:MGI Symbol;Acc:MGI:1098703] |

Retrocopy-Parental alignment summary:
>retro_mmus_621
CAGGACTGTGAGCACACCATTGGCATTGATCTCCAATACAGCCAGGCCTATGGCCTCATGGGCCTTGCGCTGTCCAGTCT
GAACAAACGTGGAGGCTGTGGTGTACTACAAGAAAACCCTGGAGATGGATTCTGACAACTACTCGTACAAGTTTAACCTC
AAGATTGCAGAGTTACAGGAGGCACCGAGCCCCACGGGTGGCATGGGCACCTTAGACATAGGTGGCTTGCTGAACAACCC
ACACTTCATCACCATGGGTTGGATCTTAATGTACAGCCCCCAACTGCAGCAGTTCATGTTGAACATGATCTCTGGAGGCC
ATAACCCTCTGGGCACTCCAGGCAGCAGTCCACAGCAGAGTGACCTGGCCAGCCTCATCCAGGTGGACCAGCAGTTTGCA
CACTAGATGGAGCAGCAGAACTCAGAATTCGTGGAGCAAATCCAGAGCCAGGTAGTACGCAGCCAGACACCCAGGGCCAG
CCATGAGGTGCAGCAAGAT
CAGGACTGTGAGCACACCATTGGCATTGATCTCCAATACAGCCAGGCCTATGGCCTCATGGGCCTTGCGCTGTCCAGTCT
GAACAAACGTGGAGGCTGTGGTGTACTACAAGAAAACCCTGGAGATGGATTCTGACAACTACTCGTACAAGTTTAACCTC
AAGATTGCAGAGTTACAGGAGGCACCGAGCCCCACGGGTGGCATGGGCACCTTAGACATAGGTGGCTTGCTGAACAACCC
ACACTTCATCACCATGGGTTGGATCTTAATGTACAGCCCCCAACTGCAGCAGTTCATGTTGAACATGATCTCTGGAGGCC
ATAACCCTCTGGGCACTCCAGGCAGCAGTCCACAGCAGAGTGACCTGGCCAGCCTCATCCAGGTGGACCAGCAGTTTGCA
CACTAGATGGAGCAGCAGAACTCAGAATTCGTGGAGCAAATCCAGAGCCAGGTAGTACGCAGCCAGACACCCAGGGCCAG
CCATGAGGTGCAGCAAGAT
ORF - retro_mmus_621 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 74.71 % |
| Parental protein coverage: | 53.65 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | QDCERAIGIDPGYSKAYGRMGLALSSLNKHA-EAVAYYKKALELDPDNDTYKSNLKIAELKLREAPSPTG |
| QDCE..IGID..YS.AYG.MGLALSSLNK.....V.Y.KK.LE.D.DN..YK.NLKIAE..L.EAPSPTG | |
| Retrocopy | QDCEHTIGIDLQYSQAYGLMGLALSSLNKRG>AVVYY-KKTLEMDSDNYSYKFNLKIAE--LQEAPSPTG |
| Parental | GVGSLDIAGLLNNPHFITMASSLMNSPQLQQLMSGMISGGHNPLGTPGSSPQQSDLASLIQAGQQFAQQM |
| G.G.LDI.GLLNNPHFITM...LM.SPQLQQ.M..MISGGHNPLGTPGSSPQQSDLASLIQ..QQFA..M | |
| Retrocopy | GMGTLDIGGLLNNPHFITMGWILMYSPQLQQFMLNMISGGHNPLGTPGSSPQQSDLASLIQVDQQFAH*M |
| Parental | QQQNPEFVEQIRSQVVRSRTPSASHEEQQE |
| .QQN.EFVEQI.SQVVRS.TP.ASHE.QQ. | |
| Retrocopy | EQQNSEFVEQIQSQVVRSQTPRASHEVQQD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .00 RPM | 79 .13 RPM |
| SRP007412_cerebellum | 0 .00 RPM | 97 .87 RPM |
| SRP007412_heart | 0 .00 RPM | 77 .55 RPM |
| SRP007412_kidney | 0 .00 RPM | 76 .42 RPM |
| SRP007412_liver | 0 .00 RPM | 50 .49 RPM |
| SRP007412_testis | 0 .00 RPM | 131 .04 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_621 was not detected
No EST(s) were mapped for retro_mmus_621 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_2009 | 145 libraries | 195 libraries | 618 libraries | 109 libraries | 5 libraries |
The graphical summary, for retro_mmus_621 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
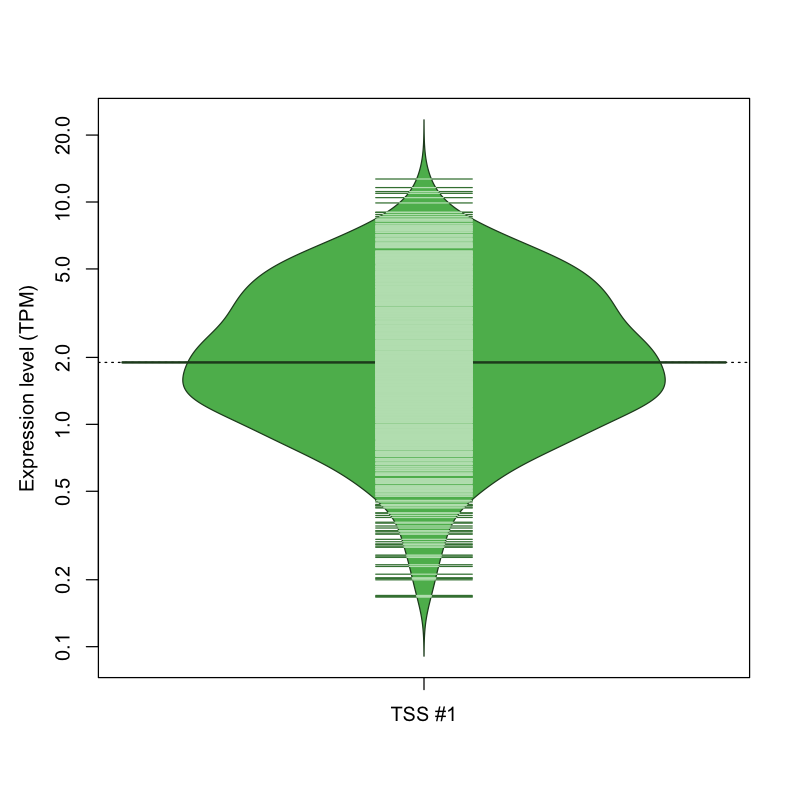
retro_mmus_621 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_621 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 5 parental genes, and 6 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000005257 | 1 retrocopy | |
| Equus caballus | ENSECAG00000010381 | 2 retrocopies | |
| Monodelphis domestica | ENSMODG00000004900 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000004937 | 1 retrocopy |
retro_mmus_621 ,
|
| Sarcophilus harrisii | ENSSHAG00000011900 | 1 retrocopy |