RetrogeneDB ID: | retro_mmus_89 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 11:35797489..35798434(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000051062 | |
| Aliases: | Fbll1, AI595406 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Fbl | ||
| Ensembl ID: | ENSMUSG00000046865 | ||
| Aliases: | Fbl, AL022665, FIB, FLRN, RNU3IP1 | ||
| Description: | fibrillarin [Source:MGI Symbol;Acc:MGI:95486] |

Retrocopy-Parental alignment summary:
>retro_mmus_89
ATGAAACCCGCGGGCGGCCGCGGCGGCTGGGGCTGGGGTGGCGGCAAGGGAGGCAGCAAGGGAGGTGACACGGGCTCAGG
GACCAAAGGCGGCTTCGGGGCACGCACACGCGGCTCCAGCGGCGGCGGCCGGGGCCGGGGACGAGGCGGCGGCGGGGGCG
GCGGGGGCGGCGGCGGCGACAGGCAGCGACGCGGCGGTCCAGGCAAGAACAAGAACCGCCGCAAGAAGGGCATCACGGTG
TCGGTGGAGCCGCATCGGCACGAGGGCGTGTTCATCTACCGCGGCGCGGAGGATGCTCTGGTCACACTGAACATGGTGCC
TGGAGTCTCGGTGTACGGCGAGAAGCGCGTCACCGTGATGGAGAACGGGGAGAAACAGGAGTACCGCACGTGGAATCCCT
TCCGCTCCAAATTGGCAGCGGCCATCCTGGGCGGCGTGGACCAGATCCACATCAAGCCCAAGTCCAAAGTGTTGTACCTG
GGCGCCGCCTCCGGAACCACTGTCTCACACGTCTCCGACATCATCGGCCCCGACGGCCTGGTCTATGCGGTTGAATTCTC
CCATCGCGCCGGCCGCGATCTGGTCAACGTGGCCAAGAAGCGCACCAACATCATCCCGGTGCTGGAAGATGCCCGGCACC
CGCTCAAGTATCGCATGCTCATTGGCATGGTGGACGTGATCTTCGCCGATGTGGCCCAGCCAGATCAGTCGCGCATCGTG
GCGCTTAACGCCCACACCTTTCTGCGCAACGGAGGCCACTTTCTCATTTCCATCAAGGCCAACTGCATCGACTCCACAGC
GTCTGCCGAGGCCGTGTTTGCTTCTGAGGTGAGGAAGCTGCAACAGGAGAACTTGAAGCCACAGGAGCAGCTGACTCTGG
AGCCCTACGAAAGGGACCACGCCGTCGTCGTCGGGGTCTATCGGCCCCCACCCAAGAGCAAGTAG
ATGAAACCCGCGGGCGGCCGCGGCGGCTGGGGCTGGGGTGGCGGCAAGGGAGGCAGCAAGGGAGGTGACACGGGCTCAGG
GACCAAAGGCGGCTTCGGGGCACGCACACGCGGCTCCAGCGGCGGCGGCCGGGGCCGGGGACGAGGCGGCGGCGGGGGCG
GCGGGGGCGGCGGCGGCGACAGGCAGCGACGCGGCGGTCCAGGCAAGAACAAGAACCGCCGCAAGAAGGGCATCACGGTG
TCGGTGGAGCCGCATCGGCACGAGGGCGTGTTCATCTACCGCGGCGCGGAGGATGCTCTGGTCACACTGAACATGGTGCC
TGGAGTCTCGGTGTACGGCGAGAAGCGCGTCACCGTGATGGAGAACGGGGAGAAACAGGAGTACCGCACGTGGAATCCCT
TCCGCTCCAAATTGGCAGCGGCCATCCTGGGCGGCGTGGACCAGATCCACATCAAGCCCAAGTCCAAAGTGTTGTACCTG
GGCGCCGCCTCCGGAACCACTGTCTCACACGTCTCCGACATCATCGGCCCCGACGGCCTGGTCTATGCGGTTGAATTCTC
CCATCGCGCCGGCCGCGATCTGGTCAACGTGGCCAAGAAGCGCACCAACATCATCCCGGTGCTGGAAGATGCCCGGCACC
CGCTCAAGTATCGCATGCTCATTGGCATGGTGGACGTGATCTTCGCCGATGTGGCCCAGCCAGATCAGTCGCGCATCGTG
GCGCTTAACGCCCACACCTTTCTGCGCAACGGAGGCCACTTTCTCATTTCCATCAAGGCCAACTGCATCGACTCCACAGC
GTCTGCCGAGGCCGTGTTTGCTTCTGAGGTGAGGAAGCTGCAACAGGAGAACTTGAAGCCACAGGAGCAGCTGACTCTGG
AGCCCTACGAAAGGGACCACGCCGTCGTCGTCGGGGTCTATCGGCCCCCACCCAAGAGCAAGTAG
ORF - retro_mmus_89 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 79.37 % |
| Parental protein coverage: | 96.02 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | GGRGGFGDRGGRGGGRGGRGGFGGGRGGFGGGGRGRGGGGGGFRGRGGGGGRGGGFQSGGNRGRGGGRG- |
| GGRGG.G..GG.GG..GG..G..G..GGFG...RG..GGG.G.RGRGGGGG.GGG...GG.R.R.GG.G. | |
| Retrocopy | GGRGGWGWGGGKGGSKGGDTG-SGTKGGFGARTRGSSGGGRG-RGRGGGGGGGGG---GGDRQRRGGPGK |
| Parental | GKRGNQSGKNVMVEPHRHEGVFICRGKEDALVTKNLVPGESVYGEKRVSISEGDDKIEYRAWNPFRSKLA |
| .K.....G..V.VEPHRHEGVFI.RG.EDALVT.N.VPG.SVYGEKRV...E...K.EYR.WNPFRSKLA | |
| Retrocopy | NKNRRKKGITVSVEPHRHEGVFIYRGAEDALVTLNMVPGVSVYGEKRVTVMENGEKQEYRTWNPFRSKLA |
| Parental | AAILGGVDQIHIKPGAKVLYLGAASGTTVSHVSDIVGPDGLVYAVEFSHRSGRDLINLAKKRTNIIPVIE |
| AAILGGVDQIHIKP..KVLYLGAASGTTVSHVSDI.GPDGLVYAVEFSHR.GRDL.N.AKKRTNIIPV.E | |
| Retrocopy | AAILGGVDQIHIKPKSKVLYLGAASGTTVSHVSDIIGPDGLVYAVEFSHRAGRDLVNVAKKRTNIIPVLE |
| Parental | DARHPHKYRMLIAMVDVIFADVAQPDQTRIVALNAHTFLRNGGHFVISIKANCIDSTASAEAVFASEVKK |
| DARHP.KYRMLI.MVDVIFADVAQPDQ.RIVALNAHTFLRNGGHF.ISIKANCIDSTASAEAVFASEV.K | |
| Retrocopy | DARHPLKYRMLIGMVDVIFADVAQPDQSRIVALNAHTFLRNGGHFLISIKANCIDSTASAEAVFASEVRK |
| Parental | MQQENMKPQEQLTLEPYERDHAVVVGVYRPPPKVK |
| .QQEN.KPQEQLTLEPYERDHAVVVGVYRPPPK.K | |
| Retrocopy | LQQENLKPQEQLTLEPYERDHAVVVGVYRPPPKSK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 13 .78 RPM | 9 .37 RPM |
| SRP007412_cerebellum | 8 .51 RPM | 12 .46 RPM |
| SRP007412_heart | 0 .00 RPM | 19 .27 RPM |
| SRP007412_kidney | 0 .00 RPM | 22 .25 RPM |
| SRP007412_liver | 0 .00 RPM | 22 .80 RPM |
| SRP007412_testis | 4 .12 RPM | 21 .31 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_89 was not detected
No EST(s) were mapped for retro_mmus_89 retrocopy.
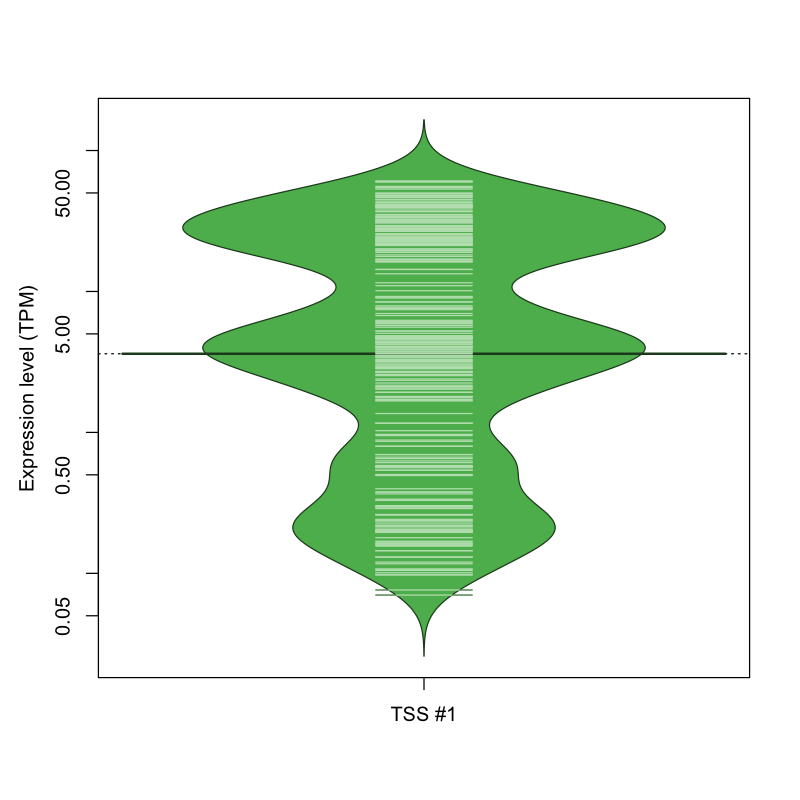
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_12351 | 813 libraries | 74 libraries | 71 libraries | 22 libraries | 92 libraries |
The graphical summary, for retro_mmus_89 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_89 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_89 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_75 |
Parental genes homology:
Parental genes homology involve 13 parental genes, and 22 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Bos taurus | ENSBTAG00000002579 | 1 retrocopy | |
| Canis familiaris | ENSCAFG00000005412 | 1 retrocopy | |
| Homo sapiens | ENSG00000105202 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000014684 | 1 retrocopy | |
| Microcebus murinus | ENSMICG00000007686 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000046865 | 4 retrocopies | |
| Nomascus leucogenys | ENSNLEG00000014234 | 1 retrocopy | |
| Otolemur garnettii | ENSOGAG00000006463 | 2 retrocopies | |
| Pan troglodytes | ENSPTRG00000010982 | 2 retrocopies | |
| Pteropus vampyrus | ENSPVAG00000010849 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000019229 | 4 retrocopies | |
| Sus scrofa | ENSSSCG00000030510 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000007755 | 2 retrocopies |