RetrogeneDB ID: | retro_hsap_4196 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 9:98908291..98910135(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000224546 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | EIF4B | ||
| Ensembl ID: | ENSG00000063046 | ||
| Aliases: | EIF4B, EIF-4B, PRO1843 | ||
| Description: | eukaryotic translation initiation factor 4B [Source:HGNC Symbol;Acc:3285] |

Retrocopy-Parental alignment summary:
>retro_hsap_4196
GCGGCCTCAGCAAAAAAGAAGAAGAAGAAGGGGAAGACTATCTCCCTAACAGACTTTCTGGCTGAGGATGGGGGTACTGG
TGGAGGAAGGACCTATGTTTCCAAACCAGTCAGCTGAGCTGATGAAACGGATGACCTGGAAGGAGATGTTTCGACAACTT
GGCACAGTAACGATGACGATGTGTATAGGGCGTCTCCAATTGACCGTTCCATCCTTCCCACTGCTCCACGGGCTGCTCGG
GAACCCAATATCGACCGGAGCCATCTTCCCAAATCGCCACCCTACACTGCTTTTCTAGGAAACCTACCCTATGATGTTAC
AGAAGAGTCAATTAAGGAATTCTTTCGAGGATTAAATATCAGTGCAGTGCGTTTACCATGTGAACCCAGCAATCCAGTGA
GGTTGAAAGGTTTTGGTTATGCTGAATTTGAGGACCTGGATTCCCTGCTTAGTGCCCTGAGTCTCAAGGAAGAGTCTCTA
GGTAACAGGAGAATTCGAGTGGACGTTGCTGATCAAGCACAGGATAAAGACAGGGATGATTGTTCTTTTGGCCGTGATAG
AAATCGGGATTCTGACAAAACAGGTACAGACTGGAGGGCTCGTCCTGCTACAGACAGCTTTGATGACTACCCACCTAGAA
GAGGTGATGATAGCTTTGGAGACAAGTATCGAGATCGTTATGATTCAGACCGGTATCGGGATGGGTATCGGGATGGGTAT
CGGGATGGCCAACGCCGGGATATGGATCCATATGGTGGCCGGGATCGCTATGATGACCGAGGCAGCAGAGACTATGATAG
AGGCTATGATTCCCGGATAGGCAGTGGCAGAAGAGCATTTGGCAGTGGGTATCGCAGGGATGATGTCTCAGAGGAGGCGG
GGACCACTATGAAGACCGATATGACAGACGGGATGATCGGTCTTGGAGCTCCAGAGATGATTACTCTCTGGATGATTATA
GGCGTGATGGTAGAGGTCCCCCCCAAAGACCCAAACTGAATCTAAACCCTCGGAGTACTCCTAAGGAAGATGATTCCTCT
GCTAGTACCTCCCAGTCCACTCGAGCTGCTTCTATCTTTGGAGGGGCAAAGCCCGTTGACACAGCTGCTAGAGAAAGAGA
AGTAGAAGAACGGCTACAGAAGGAACAAGAGAAGTTGCAGCATCAGCTGGATGAGCCAAAACTAGAACGACGGCCTCCGG
AGAGACACCCAAGCTGGCGAAGTGAAGAAACTCAGGAACGGGAACGGTCGAGGACAGGAAGTGAGTCATCACAGACTGGG
ACCTCCACCACATCTGGCAGAAGTAAGTCAGCCCAGGATGCACGAAGGAGAGAGAATGAGAAGTCTCTAGAAAATGAAAC
ACTCAATAAGGAGGAAGATTGCCACTCTCCAACTTCTAAACCTCCCAAACCTGATCAGCCCCTAAAGGTAATGCCAGCCC
CTCCACCAAAGGAGAATGCTTGGGTGAAGCGAAGTTCTAACCCTCCTGCTCGATCTCAGAGCTCAGACACAGAGCAGCAA
TCCCCTACAAGTGGTGAGGGAAAAGTAGCTCCAGCTCAACCATCTGAGGAAGGACCAGCAAGGAAAGATGAAAACAAAGT
AGATGGGATGAATGTCCCAAAAGGCCAAACTGGGAACTCTAGCCGTGGTCCAGGAGATGGAGGGAACAAAGACCACTGGA
AGGAGTCAGATAGGAAAGATGGCAAAAAGGATCAAGACTCCAGATCTGCACCTGAGCCAAAGAAACCTGAGGAAAATCCA
GCTTCCAAGTTCAGTTCTGCAAGCAAGTATGCTGCTCTCTCTGTTGATGGTGAAGATGAAAATGAGGGAGAAGATTATGC
CGAA
GCGGCCTCAGCAAAAAAGAAGAAGAAGAAGGGGAAGACTATCTCCCTAACAGACTTTCTGGCTGAGGATGGGGGTACTGG
TGGAGGAAGGACCTATGTTTCCAAACCAGTCAGCTGAGCTGATGAAACGGATGACCTGGAAGGAGATGTTTCGACAACTT
GGCACAGTAACGATGACGATGTGTATAGGGCGTCTCCAATTGACCGTTCCATCCTTCCCACTGCTCCACGGGCTGCTCGG
GAACCCAATATCGACCGGAGCCATCTTCCCAAATCGCCACCCTACACTGCTTTTCTAGGAAACCTACCCTATGATGTTAC
AGAAGAGTCAATTAAGGAATTCTTTCGAGGATTAAATATCAGTGCAGTGCGTTTACCATGTGAACCCAGCAATCCAGTGA
GGTTGAAAGGTTTTGGTTATGCTGAATTTGAGGACCTGGATTCCCTGCTTAGTGCCCTGAGTCTCAAGGAAGAGTCTCTA
GGTAACAGGAGAATTCGAGTGGACGTTGCTGATCAAGCACAGGATAAAGACAGGGATGATTGTTCTTTTGGCCGTGATAG
AAATCGGGATTCTGACAAAACAGGTACAGACTGGAGGGCTCGTCCTGCTACAGACAGCTTTGATGACTACCCACCTAGAA
GAGGTGATGATAGCTTTGGAGACAAGTATCGAGATCGTTATGATTCAGACCGGTATCGGGATGGGTATCGGGATGGGTAT
CGGGATGGCCAACGCCGGGATATGGATCCATATGGTGGCCGGGATCGCTATGATGACCGAGGCAGCAGAGACTATGATAG
AGGCTATGATTCCCGGATAGGCAGTGGCAGAAGAGCATTTGGCAGTGGGTATCGCAGGGATGATGTCTCAGAGGAGGCGG
GGACCACTATGAAGACCGATATGACAGACGGGATGATCGGTCTTGGAGCTCCAGAGATGATTACTCTCTGGATGATTATA
GGCGTGATGGTAGAGGTCCCCCCCAAAGACCCAAACTGAATCTAAACCCTCGGAGTACTCCTAAGGAAGATGATTCCTCT
GCTAGTACCTCCCAGTCCACTCGAGCTGCTTCTATCTTTGGAGGGGCAAAGCCCGTTGACACAGCTGCTAGAGAAAGAGA
AGTAGAAGAACGGCTACAGAAGGAACAAGAGAAGTTGCAGCATCAGCTGGATGAGCCAAAACTAGAACGACGGCCTCCGG
AGAGACACCCAAGCTGGCGAAGTGAAGAAACTCAGGAACGGGAACGGTCGAGGACAGGAAGTGAGTCATCACAGACTGGG
ACCTCCACCACATCTGGCAGAAGTAAGTCAGCCCAGGATGCACGAAGGAGAGAGAATGAGAAGTCTCTAGAAAATGAAAC
ACTCAATAAGGAGGAAGATTGCCACTCTCCAACTTCTAAACCTCCCAAACCTGATCAGCCCCTAAAGGTAATGCCAGCCC
CTCCACCAAAGGAGAATGCTTGGGTGAAGCGAAGTTCTAACCCTCCTGCTCGATCTCAGAGCTCAGACACAGAGCAGCAA
TCCCCTACAAGTGGTGAGGGAAAAGTAGCTCCAGCTCAACCATCTGAGGAAGGACCAGCAAGGAAAGATGAAAACAAAGT
AGATGGGATGAATGTCCCAAAAGGCCAAACTGGGAACTCTAGCCGTGGTCCAGGAGATGGAGGGAACAAAGACCACTGGA
AGGAGTCAGATAGGAAAGATGGCAAAAAGGATCAAGACTCCAGATCTGCACCTGAGCCAAAGAAACCTGAGGAAAATCCA
GCTTCCAAGTTCAGTTCTGCAAGCAAGTATGCTGCTCTCTCTGTTGATGGTGAAGATGAAAATGAGGGAGAAGATTATGC
CGAA
ORF - retro_hsap_4196 Open Reading Frame is not conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 93.34 % |
| Parental protein coverage: | 99.84 % |
| Number of stop codons detected: | 1 |
| Number of frameshifts detected | 1 |
Retrocopy - Parental Gene Alignment:
| Parental | AASAKKKNKKGKTISLTDFLAEDGGTGGGSTYVSKPVSWADETDDLEGDVSTTWHSNDDDVYRAPPIDRS |
| AASAKKK.KKGKTISLTDFLAEDGGTGGG.TYVSKPVS.ADETDDLEGDVSTTWHSNDDDVYRA.PIDRS | |
| Retrocopy | AASAKKKKKKGKTISLTDFLAEDGGTGGGRTYVSKPVS*ADETDDLEGDVSTTWHSNDDDVYRASPIDRS |
| Parental | ILPTAPRAAREPNIDRSRLPKSPPYTAFLGNLPYDVTEESIKEFFRGLNISAVRLPREPSNPERLKGFGY |
| ILPTAPRAAREPNIDRS.LPKSPPYTAFLGNLPYDVTEESIKEFFRGLNISAVRLP.EPSNP.RLKGFGY | |
| Retrocopy | ILPTAPRAAREPNIDRSHLPKSPPYTAFLGNLPYDVTEESIKEFFRGLNISAVRLPCEPSNPVRLKGFGY |
| Parental | AEFEDLDSLLSALSLNEESLGNRRIRVDVADQAQDKDRDDRSFGRDRNRDSDKTDTDWRARPATDSFDDY |
| AEFEDLDSLLSALSL.EESLGNRRIRVDVADQAQDKDRDD.SFGRDRNRDSDKT.TDWRARPATDSFDDY | |
| Retrocopy | AEFEDLDSLLSALSLKEESLGNRRIRVDVADQAQDKDRDDCSFGRDRNRDSDKTGTDWRARPATDSFDDY |
| Parental | PPRRGDDSFGDKYRDRYDSDRYRDGYRDGYRDGPRRDMDRYGGRDRYDDRGSRDYDRGYDSRIGSGRRAF |
| PPRRGDDSFGDKYRDRYDSD..............RRDMD.YGGRDRYDDRGSRDYDRGYDSRIGSGRRAF | |
| Retrocopy | PPRRGDDSFGDKYRDRYDSDXXXXXXXXXXXXXQRRDMDPYGGRDRYDDRGSRDYDRGYDSRIGSGRRAF |
| Parental | GSGYRRDDD-YRGGGDRYEDRYDRRDDRSWSSRDDYSRDDYRRDDRGPPQRPKLNLKPRSTPKEDDSSAS |
| GSGYRRDD...RGGGD.YEDRYDRRDDRSWSSRDDYS.DDYRRD.RGPPQRPKLNL.PRSTPKEDDSSAS | |
| Retrocopy | GSGYRRDDV<LRGGGDHYEDRYDRRDDRSWSSRDDYSLDDYRRDGRGPPQRPKLNLNPRSTPKEDDSSAS |
| Parental | TSQSTRAASIFGGAKPVDTAAREREVEERLQKEQEKLQRQLDEPKLERRPRERHPSWRSEETQERERSRT |
| TSQSTRAASIFGGAKPVDTAAREREVEERLQKEQEKLQ.QLDEPKLERRP.ERHPSWRSEETQERERSRT | |
| Retrocopy | TSQSTRAASIFGGAKPVDTAAREREVEERLQKEQEKLQHQLDEPKLERRPPERHPSWRSEETQERERSRT |
| Parental | GSESSQTGTSTTSSRSKSDQDARRRESEKSLENETLNKEEDCHSPTSKPPKPDQPLKVMPAPPPKENAWV |
| GSESSQTGTSTTS.RSKS.QDARRRE.EKSLENETLNKEEDCHSPTSKPPKPDQPLKVMPAPPPKENAWV | |
| Retrocopy | GSESSQTGTSTTSGRSKSAQDARRRENEKSLENETLNKEEDCHSPTSKPPKPDQPLKVMPAPPPKENAWV |
| Parental | KRSSNPPARSQSSDTEQQSPTSGGGKVAPAQPSEEGPGRKDENKVDGMNAPKGQTGNSSRGPGDGGNRDH |
| KRSSNPPARSQSSDTEQQSPTSG.GKVAPAQPSEEGP.RKDENKVDGMN.PKGQTGNSSRGPGDGGN.DH | |
| Retrocopy | KRSSNPPARSQSSDTEQQSPTSGEGKVAPAQPSEEGPARKDENKVDGMNVPKGQTGNSSRGPGDGGNKDH |
| Parental | WKESDRKDGKKDQDSRSAPEPKKPEENPASKFSSASKYAALSVDGEDENEGEDYAE |
| WKESDRKDGKKDQDSRSAPEPKKPEENPASKFSSASKYAALSVDGEDENEGEDYAE | |
| Retrocopy | WKESDRKDGKKDQDSRSAPEPKKPEENPASKFSSASKYAALSVDGEDENEGEDYAE |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 0 .20 RPM | 703 .14 RPM |
| bodymap2_adrenal | 0 .27 RPM | 434 .24 RPM |
| bodymap2_brain | 0 .05 RPM | 232 .88 RPM |
| bodymap2_breast | 0 .19 RPM | 442 .03 RPM |
| bodymap2_colon | 0 .54 RPM | 722 .93 RPM |
| bodymap2_heart | 0 .20 RPM | 297 .04 RPM |
| bodymap2_kidney | 0 .10 RPM | 206 .91 RPM |
| bodymap2_liver | 0 .17 RPM | 106 .15 RPM |
| bodymap2_lung | 0 .00 RPM | 407 .72 RPM |
| bodymap2_lymph_node | 0 .37 RPM | 454 .43 RPM |
| bodymap2_ovary | 0 .54 RPM | 1147 .94 RPM |
| bodymap2_prostate | 0 .24 RPM | 826 .01 RPM |
| bodymap2_skeletal_muscle | 1 .24 RPM | 1007 .06 RPM |
| bodymap2_testis | 0 .23 RPM | 372 .14 RPM |
| bodymap2_thyroid | 0 .21 RPM | 489 .01 RPM |
| bodymap2_white_blood_cells | 1 .14 RPM | 716 .99 RPM |
RNA Polymerase II actvity near the 5' end of retro_hsap_4196 was not detected
No EST(s) were mapped for retro_hsap_4196 retrocopy.
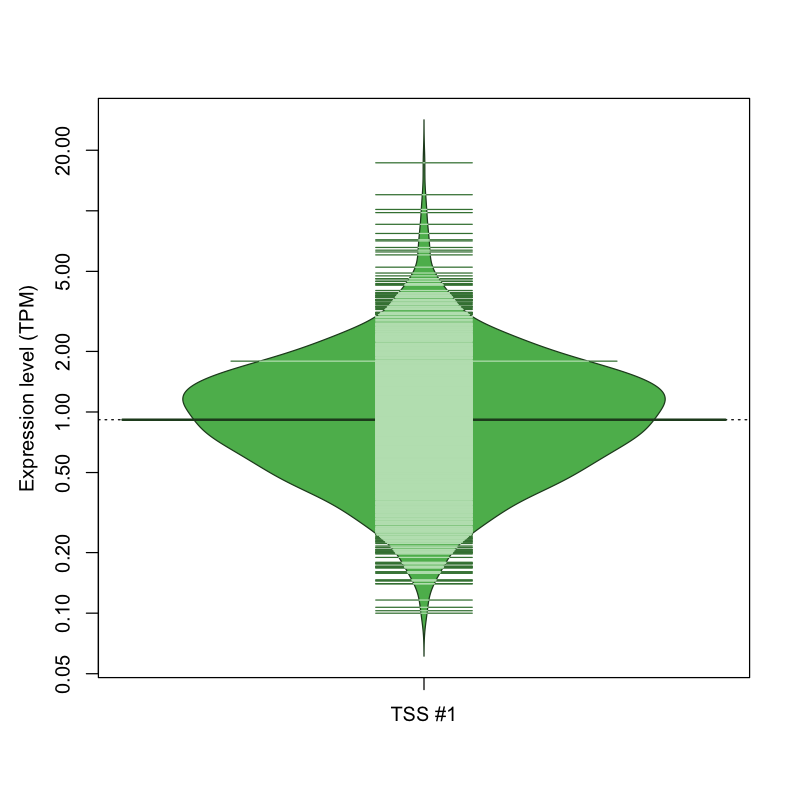
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_195254 | 434 libraries | 739 libraries | 643 libraries | 10 libraries | 3 libraries |
The graphical summary, for retro_hsap_4196 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_4196 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_4196 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 13 parental genes, and 41 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Callithrix jacchus | ENSCJAG00000018176 | 2 retrocopies | |
| Cavia porcellus | ENSCPOG00000005595 | 3 retrocopies | |
| Homo sapiens | ENSG00000063046 | 7 retrocopies |
retro_hsap_1686, retro_hsap_2639, retro_hsap_2698, retro_hsap_3767, retro_hsap_4196 , retro_hsap_4848, retro_hsap_4891,
|
| Gorilla gorilla | ENSGGOG00000013695 | 5 retrocopies | |
| Loxodonta africana | ENSLAFG00000009534 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000012341 | 2 retrocopies | |
| Macaca mulatta | ENSMMUG00000001062 | 5 retrocopies | |
| Mus musculus | ENSMUSG00000058655 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000017727 | 3 retrocopies | |
| Pongo abelii | ENSPPYG00000025848 | 7 retrocopies | |
| Rattus norvegicus | ENSRNOG00000010103 | 2 retrocopies | |
| Ictidomys tridecemlineatus | ENSSTOG00000006800 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000005423 | 2 retrocopies |
Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 4 .26 RPM |
| CEU_NA11843 | 5 .91 RPM |
| CEU_NA11930 | 6 .42 RPM |
| CEU_NA12004 | 3 .76 RPM |
| CEU_NA12400 | 6 .59 RPM |
| CEU_NA12751 | 6 .63 RPM |
| CEU_NA12760 | 5 .66 RPM |
| CEU_NA12827 | 2 .88 RPM |
| CEU_NA12872 | 3 .84 RPM |
| CEU_NA12873 | 8 .14 RPM |
| FIN_HG00183 | 3 .32 RPM |
| FIN_HG00277 | 4 .18 RPM |
| FIN_HG00315 | 6 .13 RPM |
| FIN_HG00321 | 7 .83 RPM |
| FIN_HG00328 | 6 .18 RPM |
| FIN_HG00338 | 7 .83 RPM |
| FIN_HG00349 | 5 .69 RPM |
| FIN_HG00375 | 5 .53 RPM |
| FIN_HG00377 | 3 .91 RPM |
| FIN_HG00378 | 3 .36 RPM |
| GBR_HG00099 | 6 .22 RPM |
| GBR_HG00111 | 2 .47 RPM |
| GBR_HG00114 | 4 .68 RPM |
| GBR_HG00119 | 5 .20 RPM |
| GBR_HG00131 | 6 .09 RPM |
| GBR_HG00133 | 6 .05 RPM |
| GBR_HG00134 | 3 .12 RPM |
| GBR_HG00137 | 4 .32 RPM |
| GBR_HG00142 | 3 .17 RPM |
| GBR_HG00143 | 5 .20 RPM |
| TSI_NA20512 | 4 .24 RPM |
| TSI_NA20513 | 3 .29 RPM |
| TSI_NA20518 | 4 .33 RPM |
| TSI_NA20532 | 5 .85 RPM |
| TSI_NA20538 | 5 .94 RPM |
| TSI_NA20756 | 5 .91 RPM |
| TSI_NA20765 | 5 .20 RPM |
| TSI_NA20771 | 5 .93 RPM |
| TSI_NA20786 | 4 .16 RPM |
| TSI_NA20798 | 4 .65 RPM |
| YRI_NA18870 | 5 .11 RPM |
| YRI_NA18907 | 0 .21 RPM |
| YRI_NA18916 | 0 .48 RPM |
| YRI_NA19093 | 0 .52 RPM |
| YRI_NA19099 | 0 .56 RPM |
| YRI_NA19114 | 4 .72 RPM |
| YRI_NA19118 | 0 .52 RPM |
| YRI_NA19213 | 2 .56 RPM |
| YRI_NA19214 | 0 .27 RPM |
| YRI_NA19223 | 7 .14 RPM |