RetrogeneDB ID: | retro_hsap_68 | ||
Retrocopy location | Organism: | Human (Homo sapiens) | |
| Coordinates: | 14:65007567..65009487(+) | ||
| Located in intron of: | None | ||
Retrocopy information | Ensembl ID: | ENSG00000126803 | |
| Aliases: | HSPA2, HSP70-2, HSP70-3 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental gene information | Parental gene summary: | ||
| Parental gene symbol: | HSPA8 | ||
| Ensembl ID: | ENSG00000109971 | ||
| Aliases: | HSPA8, HEL-33, HEL-S-72p, HSC54, HSC70, HSC71, HSP71, HSP73, HSPA10, LAP1, NIP71 | ||
| Description: | heat shock 70kDa protein 8 [Source:HGNC Symbol;Acc:5241] |

Retrocopy-Parental alignment summary:
>retro_hsap_68
ATGTCTGCCCGTGGCCCGGCTATCGGCATCGACCTGGGCACCACCTATTCGTGCGTCGGGGTCTTCCAACATGGCAAGGT
GGAGATCATCGCCAACGACCAGGGCAATCGCACCACCCCCAGCTACGTGGCCTTCACGGACACCGAGCGCCTCATCGGCG
ACGCCGCCAAGAACCAGGTGGCCATGAACCCCACCAACACCATCTTCGACGCCAAGAGGCTGATTGGACGGAAATTCGAG
GATGCCACAGTGCAGTCGGATATGAAACACTGGCCGTTCCGGGTGGTGAGCGAGGGAGGCAAGCCCAAAGTGCAAGTAGA
GTACAAGGGGGAGACCAAGACCTTCTTCCCAGAGGAGATATCCTCCATGGTCCTCACGAAGATGAAGGAGATCGCGGAAG
CCTACCTGGGGGGCAAGGTGCACAGCGCGGTCATAACGGTCCCGGCCTATTTCAACGACTCGCAGCGCCAGGCCACCAAG
GACGCAGGCACCATCACGGGGCTCAATGTGCTGCGCATCATCAACGAGCCCACGGCGGCGGCCATCGCCTACGGCCTGGA
CAAGAAGGGCTGCGCGGGCGGCGAGAAGAACGTGCTCATCTTTGACCTGGGCGGTGGCACTTTCGACGTGTCCATCCTGA
CCATCGAGGATGGCATCTTCGAGGTGAAGTCCACGGCCGGCGACACCCACCTGGGCGGTGAGGACTTCGACAACCGCATG
GTGAGCCACCTGGCGGAGGAGTTCAAGCGCAAGCACAAGAAGGACATTGGGCCCAACAAGCGCGCCGTGAGGCGGCTGCG
CACCGCTTGCGAGCGCGCCAAGCGCACCCTGAGCTCGTCCACGCAGGCGAGCATCGAGATCGACTCGCTCTACGAGGGCG
TGGACTTCTATACGTCCATCACGCGCGCCCGCTTCGAGGAGCTCAATGCCGACCTCTTTCGCGGGACCCTGGAGCCGGTG
GAGAAGGCGCTGCGCGACGCCAAGCTGGACAAGGGCCAGATCCAGGAGATCGTGCTGGTGGGCGGCTCCACTCGTATCCC
CAAGATCCAGAAGCTGCTGCAGGATTTCTTCAACGGCAAGGAGCTGAACAAGAGCATCAACCCCGACGAGGCGGTGGCCT
ATGGCGCCGCGGTGCAGGCGGCCATCCTCATCGGCGACAAATCAGAGAATGTGCAGGACCTGCTGCTACTCGACGTGACC
CCGTTGTCGCTGGGCATCGAGACAGCTGGCGGTGTCATGACCCCACTCATCAAGAGGAACACCACGATCCCCACCAAGCA
GACGCAGACCTTCACCACCTACTCGGACAACCAGAGCAGCGTACTGGTGCAGGTATACGAGGGCGAACGGGCCATGACCA
AGGACAATAACCTGCTGGGCAAGTTCGACCTGACCGGGATTCCCCCTGCGCCTCGCGGGGTCCCCCAAATCGAGGTTACC
TTCGACATTGACGCCAATGGCATCCTTAACGTTACCGCCGCCGACAAGAGCACCGGTAAGGAAAACAAAATCACCATCAC
CAATGACAAAGGTCGTCTGAGCAAGGACGACATTGACCGGATGGTGCAGGAGGCGGAGCGGTACAAATCGGAAGATGAGG
CGAATCGCGACCGAGTCGCGGCCAAAAACGCCCTGGAGTCCTATACCTACAACATCAAGCAGACGGTGGAAGACGAGAAA
CTGAGGGGCAAGATTAGCGAGCAGGACAAAAACAAGATCCTCGACAAGTGTCAGGAGGTGATCAACTGGCTCGACCGAAA
CCAGATGGCAGAGAAAGATGAGTATGAACACAAGCAGAAAGAGCTCGAAAGAGTTTGCAACCCCATCATCAGCAAACTTT
ACCAAGGTGGTCCTGGCGGCGGCAGCGGCGGCGGCGGTTCAGGAGCCTCCGGGGGACCCACCATCGAAGAAGTGGACTAA
ORF - retro_hsap_68 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 86.24 % |
| Parental protein coverage: | 99.85 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected: | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | SKGPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTNTVFDAK |
| ..GPA.GIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTNT.FDAK | |
| Retrocopy | ARGPAIGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPTNTIFDAK |
| Parental | RLIGRRFDDAVVQSDMKHWPFMVVNDAGRPKVQVEYKGETKSFYPEEVSSMVLTKMKEIAEAYLGKTVTN |
| RLIGR.F.DA.VQSDMKHWPF.VV...G.PKVQVEYKGETK.F.PEE.SSMVLTKMKEIAEAYLG..V.. | |
| Retrocopy | RLIGRKFEDATVQSDMKHWPFRVVSEGGKPKVQVEYKGETKTFFPEEISSMVLTKMKEIAEAYLGGKVHS |
| Parental | AVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAIAYGLDKK--VGAERNVLIFDLGGGTFDVSI |
| AV.TVPAYFNDSQRQATKDAGTI.GLNVLRIINEPTAAAIAYGLDKK...G.E.NVLIFDLGGGTFDVSI | |
| Retrocopy | AVITVPAYFNDSQRQATKDAGTITGLNVLRIINEPTAAAIAYGLDKKGCAGGEKNVLIFDLGGGTFDVSI |
| Parental | LTIEDGIFEVKSTAGDTHLGGEDFDNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQ |
| LTIEDGIFEVKSTAGDTHLGGEDFDNRMV.H...EFKRKHKKDI..NKRAVRRLRTACERAKRTLSSSTQ | |
| Retrocopy | LTIEDGIFEVKSTAGDTHLGGEDFDNRMVSHLAEEFKRKHKKDIGPNKRAVRRLRTACERAKRTLSSSTQ |
| Parental | ASIEIDSLYEGIDFYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKL |
| ASIEIDSLYEG.DFYTSITRARFEELNADLFRGTL.PVEKALRDAKLDK.QI..IVLVGGSTRIPKIQKL | |
| Retrocopy | ASIEIDSLYEGVDFYTSITRARFEELNADLFRGTLEPVEKALRDAKLDKGQIQEIVLVGGSTRIPKIQKL |
| Parental | LQDFFNGKELNKSINPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTT |
| LQDFFNGKELNKSINPDEAVAYGAAVQAAIL.GDKSENVQDLLLLDVTPLSLGIETAGGVMT.LIKRNTT | |
| Retrocopy | LQDFFNGKELNKSINPDEAVAYGAAVQAAILIGDKSENVQDLLLLDVTPLSLGIETAGGVMTPLIKRNTT |
| Parental | IPTKQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDANGILNVS |
| IPTKQTQTFTTYSDNQ..VL.QVYEGERAMTKDNNLLGKF.LTGIPPAPRGVPQIEVTFDIDANGILNV. | |
| Retrocopy | IPTKQTQTFTTYSDNQSSVLVQVYEGERAMTKDNNLLGKFDLTGIPPAPRGVPQIEVTFDIDANGILNVT |
| Parental | AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSLESYAFNMKATVEDEKLQ |
| A.DKSTGKENKITITNDKGRLSK.DI.RMVQEAE.YK.EDE..RD.V..KN.LESY..N.K.TVEDEKL. | |
| Retrocopy | AADKSTGKENKITITNDKGRLSKDDIDRMVQEAERYKSEDEANRDRVAAKNALESYTYNIKQTVEDEKLR |
| Parental | GKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELEKVCNPIITKLYQSAGGMPGGMPGGFPGGG |
| GKI...DK.KILDKC.E.INWLD.NQ.AEK.E.EH.QKELE.VCNPII.KLYQ...G.PGG..GG...GG | |
| Retrocopy | GKISEQDKNKILDKCQEVINWLDRNQMAEKDEYEHKQKELERVCNPIISKLYQ---GGPGGGSGG---GG |
| Parental | APPSGGASSGPTIEEVD |
| ...SGG....PTIEEVD | |
| Retrocopy | SGASGG----PTIEEVD |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 7 .24 RPM | 378 .65 RPM |
| bodymap2_adrenal | 20 .69 RPM | 331 .67 RPM |
| bodymap2_brain | 136 .49 RPM | 863 .93 RPM |
| bodymap2_breast | 13 .64 RPM | 569 .52 RPM |
| bodymap2_colon | 73 .44 RPM | 1201 .80 RPM |
| bodymap2_heart | 10 .39 RPM | 645 .86 RPM |
| bodymap2_kidney | 55 .90 RPM | 1396 .28 RPM |
| bodymap2_liver | 1 .37 RPM | 656 .44 RPM |
| bodymap2_lung | 14 .82 RPM | 884 .51 RPM |
| bodymap2_lymph_node | 15 .50 RPM | 1648 .41 RPM |
| bodymap2_ovary | 14 .69 RPM | 901 .10 RPM |
| bodymap2_prostate | 23 .97 RPM | 540 .08 RPM |
| bodymap2_skeletal_muscle | 7 .06 RPM | 473 .34 RPM |
| bodymap2_testis | 130 .58 RPM | 864 .58 RPM |
| bodymap2_thyroid | 23 .99 RPM | 1387 .20 RPM |
| bodymap2_white_blood_cells | 0 .69 RPM | 1500 .51 RPM |
RNA Polymerase II activity may be related with retro_hsap_68 in 40 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF002CFW | POLR2A | 14:65007304..65007764 |
| ENCFF002CFW | POLR2A | 14:65006807..65007204 |
| ENCFF002CFX | POLR2A | 14:65007304..65007758 |
| ENCFF002CFX | POLR2A | 14:65006816..65007221 |
| ENCFF002CGN | POLR2A | 14:65007339..65007641 |
| ENCFF002CGN | POLR2A | 14:65006821..65007110 |
| ENCFF002CGN | POLR2A | 14:65006131..65006595 |
| ENCFF002CHO | POLR2A | 14:65007282..65007729 |
| ENCFF002CIH | POLR2A | 14:65007432..65007571 |
| ENCFF002CIO | POLR2A | 14:65007312..65007681 |
| ENCFF002CJE | POLR2A | 14:65007248..65007757 |
| ENCFF002CJE | POLR2A | 14:65006281..65006917 |
| ENCFF002CJZ | POLR2A | 14:65006768..65007167 |
| ENCFF002CJZ | POLR2A | 14:65006389..65006939 |
| ENCFF002CKX | POLR2A | 14:65006909..65007135 |
| ENCFF002CLM | POLR2A | 14:65007201..65007741 |
| ENCFF002CLM | POLR2A | 14:65006367..65006907 |
| ENCFF002COJ | POLR2A | 14:65007228..65007664 |
| ENCFF002CPG | POLR2A | 14:65007392..65007609 |
| ENCFF002CPH | POLR2A | 14:65007277..65007667 |
| ENCFF002CQA | POLR2A | 14:65007405..65007610 |
| ENCFF002CQC | POLR2A | 14:65007315..65007669 |
| ENCFF002CQE | POLR2A | 14:65007324..65007657 |
| ENCFF002CQG | POLR2A | 14:65007348..65007614 |
| ENCFF002CQI | POLR2A | 14:65007320..65007661 |
| ENCFF002CQK | POLR2A | 14:65007308..65007676 |
| ENCFF002CQM | POLR2A | 14:65007329..65007638 |
| ENCFF002CQO | POLR2A | 14:65007354..65007671 |
| ENCFF002CRK | POLR2A | 14:65006729..65007253 |
| ENCFF002CRO | POLR2A | 14:65006749..65007169 |
| ENCFF002CRO | POLR2A | 14:65007292..65007712 |
| ENCFF002CSY | POLR2A | 14:65006747..65007234 |
| ENCFF002CSY | POLR2A | 14:65006540..65006851 |
| ENCFF002CUP | POLR2A | 14:65006805..65007121 |
| ENCFF002CUQ | POLR2A | 14:65006741..65007245 |
| ENCFF002CVF | POLR2A | 14:65007343..65007626 |
| ENCFF002CVJ | POLR2A | 14:65007257..65007705 |
| ENCFF002CZC | POLR2A | 14:65007258..65007692 |
| ENCFF002CZC | POLR2A | 14:65006780..65007143 |
| ENCFF002CZD | POLR2A | 14:65007275..65007675 |
| ENCFF002CZW | POLR2A | 14:65007225..65007770 |
| ENCFF002CZY | POLR2A | 14:65006873..65007142 |
| ENCFF002DAE | POLR2A | 14:65007377..65007622 |
| ENCFF002DAE | POLR2A | 14:65006836..65007146 |
| ENCFF002DAS | POLR2A | 14:65007237..65007713 |
| ENCFF002DAV | POLR2A | 14:65006708..65007252 |
| ENCFF002DBB | POLR2A | 14:65007387..65007587 |
| ENCFF002DBB | POLR2A | 14:65006881..65007081 |
| ENCFF002DBO | POLR2A | 14:65007346..65007650 |
| ENCFF002DBO | POLR2A | 14:65006821..65007153 |
| ENCFF002DBP | POLR2A | 14:65007353..65007635 |
| ENCFF002DBP | POLR2A | 14:65006844..65007117 |
| ENCFF002DBQ | POLR2A | 14:65007390..65007586 |
| ENCFF002DBQ | POLR2A | 14:65006873..65007065 |
| ENCFF002DBT | POLR2A | 14:65007367..65007619 |
| ENCFF002DBT | POLR2A | 14:65006777..65007147 |
200 EST(s) were mapped to retro_hsap_68 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AL044984 | 65008101 | 65008703 | 99.9 | 601 | 1 | 600 |
| AL046354 | 65007688 | 65008189 | 100 | 501 | 0 | 501 |
| BE894081 | 65007867 | 65008582 | 99.1 | 700 | 1 | 688 |
| BE894894 | 65007501 | 65008190 | 98.1 | 680 | 0 | 668 |
| BF969405 | 65007779 | 65008539 | 99.7 | 751 | 1 | 743 |
| BG037156 | 65007463 | 65008150 | 99.1 | 658 | 5 | 643 |
| BG331894 | 65007722 | 65008374 | 96.1 | 628 | 7 | 609 |
| BG476580 | 65007478 | 65008272 | 95.2 | 770 | 6 | 738 |
| BG699527 | 65008101 | 65008739 | 99.9 | 635 | 1 | 632 |
| BG701913 | 65007462 | 65008145 | 98.6 | 678 | 4 | 670 |
| BG704017 | 65007462 | 65008296 | 98.7 | 807 | 4 | 788 |
| BG716913 | 65007466 | 65008076 | 100 | 610 | 0 | 610 |
| BG717819 | 65007458 | 65008156 | 99.8 | 695 | 1 | 691 |
| BG724346 | 65007462 | 65008137 | 97.2 | 669 | 5 | 656 |
| BG759101 | 65007830 | 65008479 | 99.9 | 648 | 1 | 647 |
| BG771669 | 65007462 | 65008235 | 97.8 | 767 | 3 | 753 |
| BG772001 | 65007462 | 65008261 | 99.5 | 784 | 2 | 776 |
| BG772257 | 65007462 | 65008211 | 97 | 738 | 5 | 718 |
| BG772613 | 65007462 | 65008118 | 98.1 | 653 | 3 | 645 |
| BI085619 | 65008000 | 65008812 | 97.8 | 809 | 3 | 798 |
| BI094536 | 65007595 | 65008539 | 98.9 | 686 | 3 | 674 |
| BI463605 | 65007459 | 65008266 | 98.5 | 789 | 2 | 774 |
| BI544491 | 65007448 | 65008299 | 96.8 | 834 | 5 | 809 |
| BI550887 | 65007457 | 65008098 | 99.9 | 638 | 1 | 635 |
| BI560802 | 65007462 | 65008342 | 96.1 | 782 | 9 | 757 |
| BI562664 | 65007448 | 65008254 | 97.1 | 789 | 3 | 769 |
| BI596909 | 65007448 | 65008209 | 99 | 752 | 3 | 743 |
| BI601270 | 65007448 | 65008233 | 98.8 | 763 | 9 | 745 |
| BI601546 | 65007394 | 65008302 | 96.8 | 821 | 8 | 790 |
| BI604199 | 65007459 | 65008099 | 97.4 | 635 | 5 | 624 |
| BI667585 | 65007515 | 65008264 | 97.5 | 732 | 3 | 717 |
| BI668472 | 65007463 | 65008153 | 97.6 | 686 | 3 | 674 |
| BI761630 | 65007481 | 65008067 | 100 | 586 | 0 | 586 |
| BI761806 | 65007475 | 65008229 | 98.6 | 751 | 1 | 743 |
| BM017695 | 65007956 | 65008770 | 97.3 | 810 | 3 | 794 |
| BM453285 | 65007498 | 65008262 | 98.9 | 759 | 4 | 753 |
| BM471796 | 65007468 | 65008266 | 98.3 | 791 | 7 | 781 |
| BM706658 | 65007478 | 65008100 | 100 | 622 | 0 | 622 |
| BM905244 | 65007475 | 65008290 | 98.4 | 792 | 3 | 782 |
| BP214223 | 65007458 | 65008046 | 99.9 | 587 | 1 | 586 |
| BP214389 | 65007458 | 65008511 | 98.9 | 590 | 5 | 582 |
| BP214418 | 65007458 | 65008057 | 99.7 | 597 | 2 | 595 |
| BP214723 | 65007459 | 65008042 | 100 | 583 | 0 | 583 |
| BP215418 | 65007460 | 65008042 | 99.9 | 581 | 1 | 580 |
| BP216662 | 65007461 | 65008044 | 99.9 | 582 | 1 | 581 |
| BP280320 | 65007463 | 65008051 | 100 | 588 | 0 | 588 |
| BP320268 | 65007459 | 65008047 | 100 | 588 | 0 | 588 |
| BP360968 | 65007458 | 65008046 | 99.5 | 585 | 3 | 582 |
| BP371837 | 65008218 | 65008800 | 99.9 | 581 | 1 | 580 |
| BP374622 | 65007460 | 65008043 | 100 | 583 | 0 | 583 |
| BQ232549 | 65007553 | 65008435 | 97.3 | 855 | 7 | 832 |
| BQ339091 | 65008029 | 65008601 | 99.7 | 567 | 2 | 562 |
| BQ431623 | 65007533 | 65008353 | 97.8 | 812 | 2 | 799 |
| BQ435120 | 65007481 | 65008246 | 99.3 | 761 | 0 | 758 |
| BQ441040 | 65007484 | 65008321 | 95.8 | 826 | 5 | 798 |
| BQ670728 | 65007468 | 65008212 | 99.5 | 720 | 3 | 714 |
| BQ676345 | 65007857 | 65008572 | 99.3 | 709 | 5 | 703 |
| BQ719746 | 65007486 | 65008161 | 97.5 | 673 | 0 | 662 |
| BU161469 | 65007485 | 65008181 | 98.4 | 655 | 5 | 647 |
| BU167405 | 65007488 | 65008190 | 97.1 | 690 | 12 | 674 |
| BU182828 | 65007553 | 65008145 | 97.3 | 585 | 2 | 575 |
| BU187966 | 65007959 | 65008785 | 99 | 817 | 3 | 810 |
| BU188162 | 65007487 | 65008283 | 96.6 | 773 | 11 | 749 |
| BU188598 | 65007485 | 65008343 | 98.6 | 849 | 6 | 841 |
| BX338028 | 65007482 | 65008480 | 98.7 | 958 | 8 | 939 |
| BX359218 | 65007494 | 65008453 | 99 | 939 | 9 | 926 |
| BX359765 | 65007479 | 65008302 | 99.8 | 814 | 2 | 810 |
| BX360652 | 65007476 | 65008406 | 99.5 | 916 | 4 | 909 |
| BX381080 | 65007479 | 65008528 | 98.6 | 1026 | 12 | 1013 |
| BX428790 | 65007470 | 65008406 | 99.3 | 910 | 3 | 899 |
| BX450649 | 65008091 | 65008848 | 98.3 | 749 | 6 | 737 |
| BX459030 | 65007470 | 65008241 | 100 | 762 | 0 | 762 |
| CA392607 | 65007866 | 65008514 | 99.9 | 647 | 1 | 646 |
| CB152018 | 65007458 | 65008097 | 100 | 638 | 0 | 638 |
| CB155424 | 65007458 | 65008045 | 100 | 587 | 0 | 587 |
| CB155883 | 65007530 | 65008119 | 99.9 | 588 | 1 | 587 |
| CB215388 | 65007475 | 65008107 | 100 | 632 | 0 | 632 |
| CB988234 | 65007458 | 65008149 | 96.7 | 679 | 0 | 662 |
| CD251862 | 65007490 | 65008298 | 98.9 | 806 | 2 | 801 |
| CD252010 | 65007486 | 65008346 | 97.3 | 833 | 8 | 814 |
| CD299958 | 65007471 | 65008298 | 98 | 811 | 2 | 799 |
| CD300159 | 65007795 | 65008608 | 98.4 | 804 | 3 | 793 |
| CD300195 | 65007847 | 65008701 | 99 | 843 | 2 | 834 |
| CD300438 | 65007724 | 65008555 | 97.6 | 823 | 7 | 808 |
| CD358704 | 65007904 | 65008520 | 99.1 | 613 | 2 | 608 |
| CD514561 | 65007483 | 65008396 | 99.7 | 862 | 0 | 858 |
| CD653825 | 65007510 | 65008272 | 99.7 | 761 | 0 | 760 |
| CN356508 | 65007751 | 65008227 | 100 | 476 | 0 | 476 |
| CV024696 | 65007567 | 65008205 | 99.6 | 635 | 3 | 632 |
| CV025662 | 65007567 | 65008169 | 99.7 | 600 | 2 | 598 |
| DA057147 | 65007458 | 65008066 | 99.9 | 607 | 1 | 606 |
| DA075637 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA133516 | 65007863 | 65008428 | 99.3 | 564 | 1 | 562 |
| DA139262 | 65007458 | 65008082 | 100 | 624 | 0 | 624 |
| DA139403 | 65007458 | 65008077 | 100 | 619 | 0 | 619 |
| DA139409 | 65007458 | 65008076 | 100 | 618 | 0 | 618 |
| DA140888 | 65007458 | 65008082 | 100 | 624 | 0 | 624 |
| DA145059 | 65007459 | 65008045 | 100 | 586 | 0 | 586 |
| DA149843 | 65007458 | 65008051 | 100 | 593 | 0 | 593 |
| DA154252 | 65007458 | 65008047 | 100 | 589 | 0 | 589 |
| DA201491 | 65007458 | 65008047 | 100 | 589 | 0 | 589 |
| DA201581 | 65007458 | 65008053 | 99.7 | 593 | 2 | 591 |
| DA201703 | 65007458 | 65008451 | 100 | 634 | 0 | 633 |
| DA201747 | 65007458 | 65008083 | 99.6 | 625 | 0 | 624 |
| DA201797 | 65007458 | 65008081 | 100 | 623 | 0 | 623 |
| DA202671 | 65008015 | 65008597 | 99.7 | 579 | 2 | 576 |
| DA204759 | 65007458 | 65008046 | 99.9 | 587 | 1 | 586 |
| DA205777 | 65007458 | 65008051 | 100 | 593 | 0 | 593 |
| DA240351 | 65007458 | 65008064 | 100 | 606 | 0 | 606 |
| DA253528 | 65007458 | 65008042 | 100 | 584 | 0 | 584 |
| DA254005 | 65007458 | 65008048 | 99.9 | 589 | 1 | 588 |
| DA255673 | 65007615 | 65008133 | 100 | 518 | 0 | 518 |
| DA260197 | 65007802 | 65008372 | 99.9 | 569 | 1 | 568 |
| DA265713 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA269471 | 65007458 | 65008100 | 99.9 | 641 | 1 | 640 |
| DA279428 | 65007458 | 65008047 | 99.9 | 588 | 1 | 587 |
| DA281221 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA286463 | 65007667 | 65008229 | 100 | 562 | 0 | 562 |
| DA287417 | 65007458 | 65008043 | 100 | 585 | 0 | 585 |
| DA287480 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA290668 | 65007458 | 65008045 | 100 | 587 | 0 | 587 |
| DA293759 | 65007458 | 65008043 | 100 | 585 | 0 | 585 |
| DA298240 | 65007458 | 65008052 | 100 | 594 | 0 | 594 |
| DA300093 | 65007459 | 65008045 | 100 | 586 | 0 | 586 |
| DA301795 | 65007458 | 65008046 | 100 | 588 | 0 | 588 |
| DA301894 | 65007458 | 65008047 | 100 | 589 | 0 | 589 |
| DA302296 | 65007460 | 65008051 | 100 | 591 | 0 | 591 |
| DA303437 | 65007458 | 65008048 | 100 | 590 | 0 | 590 |
| DA303535 | 65007457 | 65008048 | 99.9 | 590 | 1 | 589 |
| DA305259 | 65007461 | 65008041 | 100 | 580 | 0 | 580 |
| DA307036 | 65007458 | 65008041 | 100 | 583 | 0 | 583 |
| DA307510 | 65007458 | 65008051 | 100 | 593 | 0 | 593 |
| DA307570 | 65007458 | 65008056 | 100 | 598 | 0 | 598 |
| DA308123 | 65007458 | 65008046 | 100 | 587 | 0 | 587 |
| DA308334 | 65007458 | 65008052 | 99.9 | 593 | 1 | 592 |
| DA309690 | 65007459 | 65008065 | 100 | 606 | 0 | 606 |
| DA342393 | 65007770 | 65008337 | 99.9 | 566 | 1 | 565 |
| DA343139 | 65007458 | 65008046 | 100 | 588 | 0 | 588 |
| DA343159 | 65007458 | 65008052 | 100 | 594 | 0 | 594 |
| DA343196 | 65007458 | 65008049 | 99.9 | 590 | 1 | 589 |
| DA346329 | 65007458 | 65008051 | 100 | 593 | 0 | 593 |
| DA349811 | 65007459 | 65008046 | 100 | 587 | 0 | 587 |
| DA353020 | 65007458 | 65008042 | 100 | 584 | 0 | 584 |
| DA354233 | 65007458 | 65008043 | 100 | 585 | 0 | 585 |
| DA354277 | 65007460 | 65008053 | 99.9 | 591 | 1 | 590 |
| DA384728 | 65007458 | 65008047 | 100 | 589 | 0 | 589 |
| DA386304 | 65007458 | 65008052 | 100 | 594 | 0 | 594 |
| DA386337 | 65007458 | 65008041 | 100 | 583 | 0 | 583 |
| DA387058 | 65007458 | 65008065 | 100 | 607 | 0 | 607 |
| DA388049 | 65007458 | 65008050 | 100 | 592 | 0 | 592 |
| DA388472 | 65007458 | 65008056 | 100 | 598 | 0 | 598 |
| DA391676 | 65007458 | 65008048 | 100 | 590 | 0 | 590 |
| DA391740 | 65007458 | 65008046 | 99.9 | 587 | 1 | 586 |
| DA391847 | 65007458 | 65008042 | 100 | 584 | 0 | 584 |
| DA392131 | 65007458 | 65008056 | 100 | 598 | 0 | 598 |
| DA392251 | 65007458 | 65008045 | 99.9 | 586 | 1 | 585 |
| DA392361 | 65007458 | 65008049 | 99.9 | 590 | 1 | 589 |
| DA394394 | 65007454 | 65008044 | 100 | 590 | 0 | 590 |
| DA394513 | 65007458 | 65008051 | 100 | 593 | 0 | 593 |
| DA394616 | 65007458 | 65008057 | 100 | 599 | 0 | 599 |
| DA395250 | 65007458 | 65008055 | 100 | 597 | 0 | 597 |
| DA397431 | 65007458 | 65008052 | 99.9 | 593 | 1 | 592 |
| DA397692 | 65007458 | 65008061 | 99.6 | 600 | 2 | 596 |
| DA397859 | 65007460 | 65008053 | 99.9 | 592 | 1 | 591 |
| DA397967 | 65007458 | 65008055 | 100 | 597 | 0 | 597 |
| DA435150 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA441135 | 65007458 | 65008041 | 100 | 583 | 0 | 583 |
| DA581031 | 65007458 | 65008210 | 97.6 | 744 | 4 | 730 |
| DA619893 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA634334 | 65007458 | 65008065 | 100 | 607 | 0 | 607 |
| DA635825 | 65007460 | 65008049 | 99.9 | 588 | 1 | 587 |
| DA846773 | 65007458 | 65008043 | 100 | 585 | 0 | 585 |
| DA847720 | 65008341 | 65008917 | 100 | 576 | 0 | 576 |
| DA936518 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DA943036 | 65007458 | 65008054 | 100 | 596 | 0 | 596 |
| DB019609 | 65007458 | 65008130 | 99.5 | 669 | 0 | 666 |
| DB027553 | 65007458 | 65008055 | 99.9 | 596 | 1 | 595 |
| DB028976 | 65007502 | 65008052 | 100 | 550 | 0 | 550 |
| DB029145 | 65007458 | 65008058 | 99.5 | 600 | 0 | 599 |
| DB030746 | 65007458 | 65008046 | 100 | 588 | 0 | 588 |
| DB030952 | 65007932 | 65008481 | 98.8 | 546 | 2 | 542 |
| DB031327 | 65007458 | 65008067 | 99.9 | 608 | 1 | 607 |
| DB033795 | 65007458 | 65008049 | 100 | 591 | 0 | 591 |
| DB034804 | 65007588 | 65008168 | 100 | 580 | 0 | 580 |
| DB037666 | 65007459 | 65008041 | 99.9 | 581 | 1 | 580 |
| DB044202 | 65007564 | 65008114 | 99.9 | 549 | 1 | 548 |
| DB167291 | 65007463 | 65008045 | 100 | 582 | 0 | 582 |
| DB168716 | 65007458 | 65008061 | 100 | 603 | 0 | 603 |
| DB171109 | 65007458 | 65008044 | 100 | 586 | 0 | 586 |
| DB171885 | 65007458 | 65008051 | 99.9 | 592 | 1 | 591 |
| DB172652 | 65007458 | 65008052 | 99.7 | 592 | 2 | 590 |
| DB247214 | 65007458 | 65008138 | 98.3 | 674 | 1 | 665 |
| DB249257 | 65007916 | 65008412 | 100 | 496 | 0 | 496 |
| DB251872 | 65007458 | 65008088 | 100 | 630 | 0 | 630 |
| DB263242 | 65007458 | 65008058 | 99.9 | 598 | 1 | 597 |
| DN991280 | 65007474 | 65008045 | 100 | 571 | 0 | 571 |
| HY008431 | 65007564 | 65008102 | 100 | 537 | 0 | 537 |
| HY011039 | 65007918 | 65008434 | 99.9 | 515 | 1 | 514 |
| HY130191 | 65008258 | 65008738 | 100 | 479 | 0 | 479 |
| HY335166 | 65008580 | 65009064 | 99.6 | 482 | 2 | 480 |
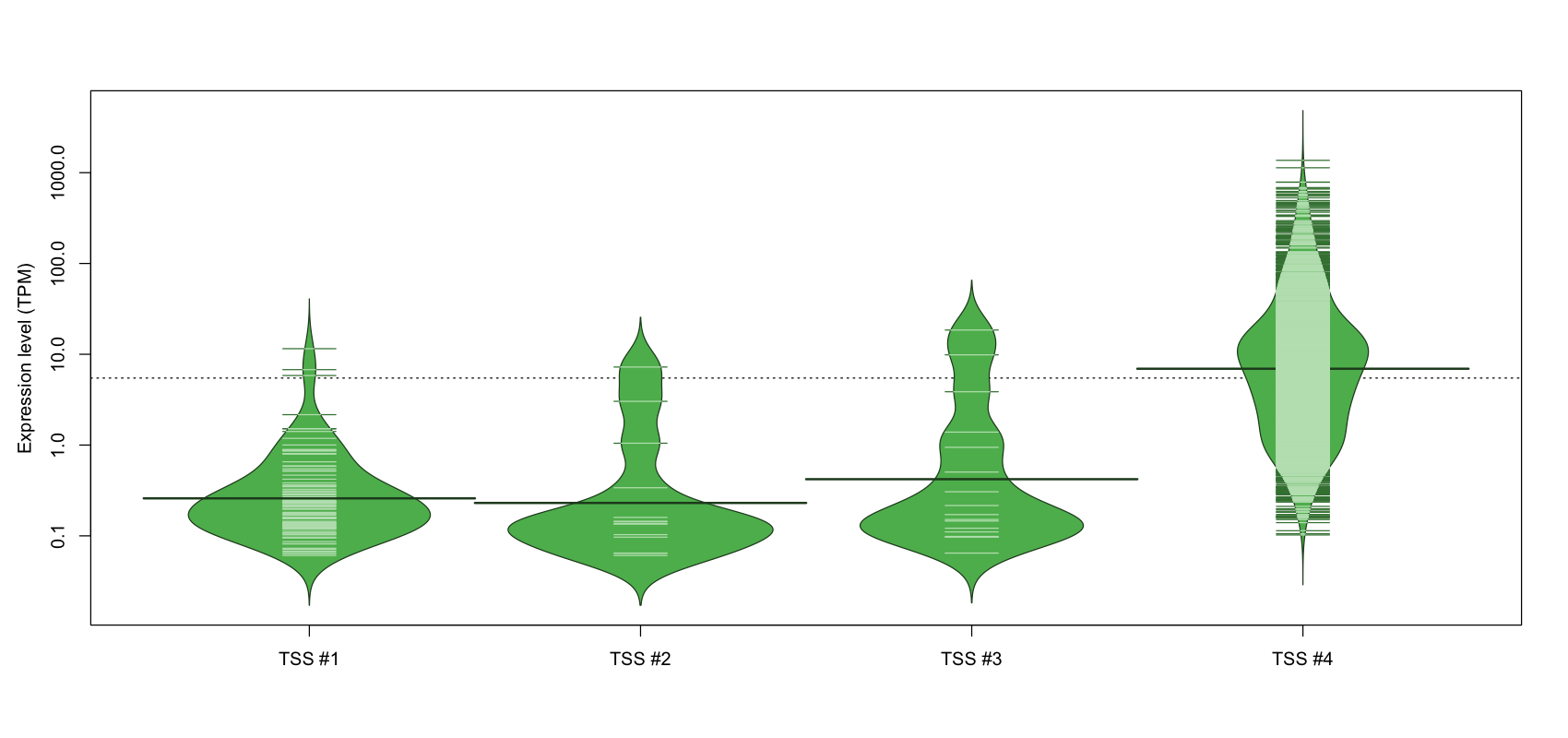
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_39560 | 1733 libraries | 87 libraries | 6 libraries | 2 libraries | 1 library |
| TSS #2 | TSS_39561 | 1815 libraries | 11 libraries | 2 libraries | 1 library | 0 libraries |
| TSS #3 | TSS_39562 | 1813 libraries | 12 libraries | 2 libraries | 1 library | 1 library |
| TSS #4 | TSS_39563 | 230 libraries | 225 libraries | 450 libraries | 259 libraries | 665 libraries |
The graphical summary, for retro_hsap_68 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_68 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_68 has 4 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Gorilla gorilla | retro_ggor_1053 |
| Pongo abelii | retro_pabe_183 |
| Bos taurus | retro_btau_354 |
| Canis familiaris | retro_cfam_140 |
Parental genes homology:
Parental genes homology involve 29 parental genes, and 76 retrocopies.Expression level across human populations :
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2089
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2112
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2135
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2158
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2181
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2204
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2227
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2250
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2273
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2296
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2325
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2348
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2371
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2394
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2417
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2440
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2463
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2486
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2509
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2532
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2553
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2576
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2599
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2622
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2645
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2668
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2691
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2714
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2737
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2760
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2787
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2810
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2837
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2860
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2887
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2910
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2937
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2960
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 2987
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3010
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3122
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3145
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3168
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3191
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3214
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3241
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3264
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3287
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3310
Warning: Undefined variable $populationsDictCols in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Trying to access array offset on value of type null in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3333
Warning: Undefined variable $populLEGEND in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3353
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 0 .00 RPM |
| CEU_NA11843 | 0 .00 RPM |
| CEU_NA11930 | 0 .00 RPM |
| CEU_NA12004 | 0 .00 RPM |
| CEU_NA12400 | 0 .00 RPM |
| CEU_NA12751 | 0 .00 RPM |
| CEU_NA12760 | 0 .00 RPM |
| CEU_NA12827 | 0 .00 RPM |
| CEU_NA12872 | 0 .00 RPM |
| CEU_NA12873 | 0 .00 RPM |
| FIN_HG00183 | 0 .00 RPM |
| FIN_HG00277 | 0 .00 RPM |
| FIN_HG00315 | 0 .00 RPM |
| FIN_HG00321 | 0 .00 RPM |
| FIN_HG00328 | 0 .00 RPM |
| FIN_HG00338 | 0 .00 RPM |
| FIN_HG00349 | 0 .00 RPM |
| FIN_HG00375 | 0 .00 RPM |
| FIN_HG00377 | 0 .00 RPM |
| FIN_HG00378 | 0 .00 RPM |
| GBR_HG00099 | 0 .00 RPM |
| GBR_HG00111 | 0 .00 RPM |
| GBR_HG00114 | 0 .00 RPM |
| GBR_HG00119 | 0 .00 RPM |
| GBR_HG00131 | 0 .00 RPM |
| GBR_HG00133 | 0 .00 RPM |
| GBR_HG00134 | 0 .00 RPM |
| GBR_HG00137 | 0 .00 RPM |
| GBR_HG00142 | 0 .00 RPM |
| GBR_HG00143 | 0 .00 RPM |
| TSI_NA20512 | 0 .00 RPM |
| TSI_NA20513 | 0 .00 RPM |
| TSI_NA20518 | 0 .00 RPM |
| TSI_NA20532 | 0 .00 RPM |
| TSI_NA20538 | 0 .00 RPM |
| TSI_NA20756 | 0 .00 RPM |
| TSI_NA20765 | 0 .00 RPM |
| TSI_NA20771 | 0 .00 RPM |
| TSI_NA20786 | 0 .00 RPM |
| TSI_NA20798 | 0 .00 RPM |
| YRI_NA18870 | 0 .00 RPM |
| YRI_NA18907 | 0 .00 RPM |
| YRI_NA18916 | 0 .00 RPM |
| YRI_NA19093 | 0 .00 RPM |
| YRI_NA19099 | 0 .00 RPM |
| YRI_NA19114 | 0 .00 RPM |
| YRI_NA19118 | 0 .00 RPM |
| YRI_NA19213 | 0 .00 RPM |
| YRI_NA19214 | 0 .00 RPM |
| YRI_NA19223 | 0 .00 RPM |
Fatal error: Uncaught Error: Call to undefined function mysql_query() in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php:3390 Stack trace: #0 /home/rosikiewicz/RetrogeneDB2/BETA/retrogene.php(1158): include() #1 {main} thrown in /home/rosikiewicz/RetrogeneDB2/BETA/retrogene_maps.php on line 3390