RetrogeneDB ID: | retro_mmus_2545 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 5:8971947..8972139(+) | ||
| Located in intron of: | ENSMUSG00000003623 | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000078183 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Tma7 | ||
| Ensembl ID: | ENSMUSG00000091537 | ||
| Aliases: | Tma7, 1110017O22Rik, Ccdc72 | ||
| Description: | translational machinery associated 7 homolog (S. cerevisiae) [Source:MGI Symbol;Acc:MGI:1913417] |

Retrocopy-Parental alignment summary:
>retro_mmus_2545
ATGTCGGGCTGGGAAGGTGGCAAAAAGAAGCCCCTGAAACAGCCCAAGAAGCAGGCCAAGGAGATGGACGAGGAAGATAA
GGCTTTCAAGCAGAATCAAAAGGAGGAACGGAAGAAACTCGAGGAGCTAAAAGCCAAGGCCGCCGGGAAGGGCCCCCTGG
CCACAGGTGGAATTAAGAAATCTGGCAAAAAG
ATGTCGGGCTGGGAAGGTGGCAAAAAGAAGCCCCTGAAACAGCCCAAGAAGCAGGCCAAGGAGATGGACGAGGAAGATAA
GGCTTTCAAGCAGAATCAAAAGGAGGAACGGAAGAAACTCGAGGAGCTAAAAGCCAAGGCCGCCGGGAAGGGCCCCCTGG
CCACAGGTGGAATTAAGAAATCTGGCAAAAAG
ORF - retro_mmus_2545 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 95.31 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSGREGGKKKPLKQPKKQAKEMDEEDKAFKQKQKEEQKKLEELKAKAAGKGPLATGGIKKSGKK |
| MSG.EGGKKKPLKQPKKQAKEMDEEDKAFKQ.QKEE.KKLEELKAKAAGKGPLATGGIKKSGKK | |
| Retrocopy | MSGWEGGKKKPLKQPKKQAKEMDEEDKAFKQNQKEERKKLEELKAKAAGKGPLATGGIKKSGKK |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .12 RPM | 10 .07 RPM |
| SRP007412_cerebellum | 0 .17 RPM | 7 .08 RPM |
| SRP007412_heart | 0 .00 RPM | 17 .53 RPM |
| SRP007412_kidney | 0 .33 RPM | 15 .34 RPM |
| SRP007412_liver | 0 .25 RPM | 11 .03 RPM |
| SRP007412_testis | 0 .69 RPM | 15 .14 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_2545 was not detected
No EST(s) were mapped for retro_mmus_2545 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_116072 | 532 libraries | 238 libraries | 276 libraries | 24 libraries | 2 libraries |
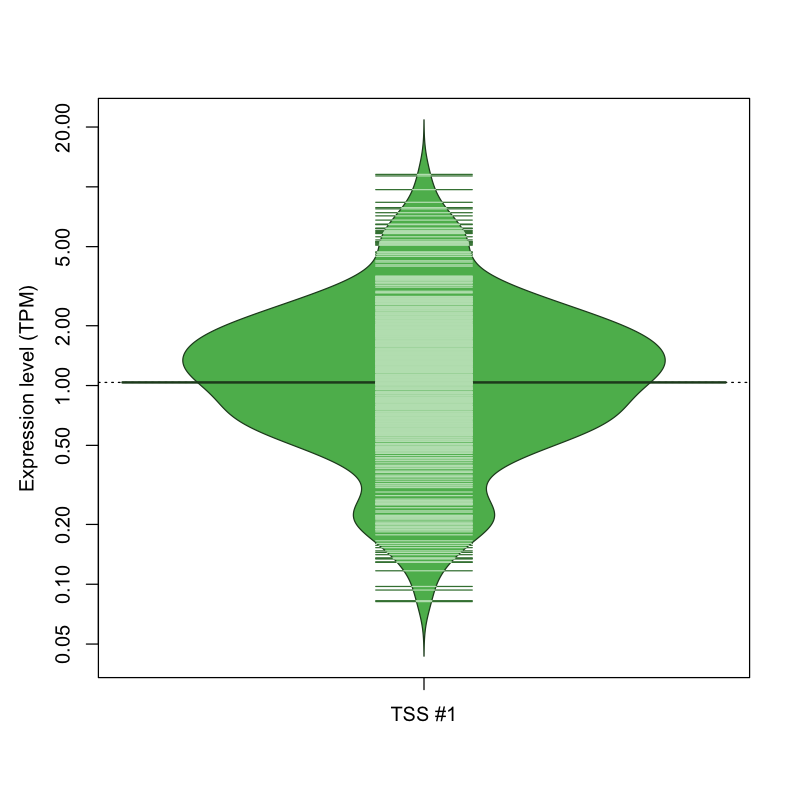
The graphical summary, for retro_mmus_2545 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_2545 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_2545 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 9 parental genes, and 61 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Ailuropoda melanoleuca | ENSAMEG00000001193 | 6 retrocopies | |
| Callithrix jacchus | ENSCJAG00000002871 | 18 retrocopies |
retro_cjac_1370, retro_cjac_1449, retro_cjac_1504, retro_cjac_1667, retro_cjac_1670, retro_cjac_1704, retro_cjac_1918, retro_cjac_2092, retro_cjac_2258, retro_cjac_2422, retro_cjac_2629, retro_cjac_2673, retro_cjac_2692, retro_cjac_2799, retro_cjac_3651, retro_cjac_536, retro_cjac_81, retro_cjac_849,
|
| Felis catus | ENSFCAG00000026902 | 5 retrocopies | |
| Gorilla gorilla | ENSGGOG00000028044 | 3 retrocopies | |
| Mus musculus | ENSMUSG00000091537 | 11 retrocopies |
retro_mmus_1216, retro_mmus_1582, retro_mmus_1632, retro_mmus_2117, retro_mmus_2189, retro_mmus_2222, retro_mmus_2545 , retro_mmus_2975, retro_mmus_3235, retro_mmus_389, retro_mmus_412,
|
| Pan troglodytes | ENSPTRG00000041769 | 9 retrocopies | |
| Rattus norvegicus | ENSRNOG00000042689 | 2 retrocopies | |
| Rattus norvegicus | ENSRNOG00000047225 | 1 retrocopy | |
| Sus scrofa | ENSSSCG00000011351 | 6 retrocopies |