RetrogeneDB ID: | retro_mmus_788 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 11:48751869..48753714(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000082454 | |
| Aliases: | None | ||
| Status: | KNOWN_PSEUDOGENE | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Ddx5 | ||
| Ensembl ID: | ENSMUSG00000020719 | ||
| Aliases: | Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, p68 | ||
| Description: | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 [Source:MGI Symbol;Acc:MGI:105037] |

Retrocopy-Parental alignment summary:
>retro_mmus_788
ATGTCGAGTTATTCTAGTGACCGAGACCACAGCCGGGATCGAGGGTTTGGTGCACCTCGATTTGGAGGGAGTAGAACAGG
ACCCCTCTCTGGAAAGAAGTTTGGAAATCCTGGGGAGAAACTAGTTAAAAAGAAGTGGAATCTTGATGAGCTGCCCAAAT
TTGAGAAGAATTTTTATCAAGAACACCCTGATTTGGCAAGGCGCACCGCACAAGAGGTTGATACATACAGAAGAAGCAAG
GAAATTACAGTTAGAGGTCACAACTGTCCAAAACCTGTTCTGAAGTTTTATGAAGCAAACTTTCCTGCGAATGTCATGGA
CGTGATTGCAAGGCAGAACTTCACTGAACCCACTGCTATTCAAGCTCAGGGCTGGCCAGTTGCTCTCAGTGGATTGGATA
TGGTTGGAGTAGCTCAGACTGGATCTGGGAAAACACTATCTTATTTGCTGCCTGCCATTGTACACATAATCCACCAGCCA
TTCCTAGAGAGAGGTGATGGGCCTATTTGCTTGGTGCTGGCACCAACTCGAGAACTGGCACAGCAGGTGCAGCAAGTGGC
TGCTGAATATTGTAGAGCTTGTCGCTTGAAGTCTACTTGCATCTATGGTGGTGCTCCCAAAGGACCACAGATTCGTGATT
TGGAAAGAGGCGTGGAAATCTGTATTGCAACACCTGGAAGACTGATTGACTTTTTAGAGTGTGGGAAAACCAATCTGAGA
AGAACAACGTACCTTGTCCTCGATGAAGCTGATAGGATGCTTGATATGGGATTTGAACCCCAGATAAGGAAAATCGTGGA
TCAAATAAGACCTGATAGGCAAACACTAATGTGGAGTGCAACTTGGCCAAAAGAAGTAAGACAGCTTGCTGAAGATTTCC
TGAAAGACTATATTCATATCAATATTGGTGCGCTGGAACTGAGTGCAAACCATAACATTCTCCAGATTGTGGATGTATGT
CATGATGTCGAAAAGGATGAAAAGCTTATTCTTCTGATGGAAGAAATCATGAGTGAGAAGGAGAATAAAACCATTGTTTT
TGTTGAAACCAAAAGAAGATGTGATGAACTTACCAGAAAAATGAGGAGAGATGGGTGGCCTGCCATGGGCATCCATGGTG
ACAAGAGTCAGCAGGAACGTGACTGGGTTCTAAGTGAATTCAAACATGGAAAAGCTTCTATTCTGATTGCTACTGATGTG
GCCTCCAGAGGGCTAGATGTGGAAGATGTGAAATTTGTCATCAATTATGACTACCCTAACTCCTCAGAGGATTATATTCA
TCGAATTGGAAGAACTGCTCGCAGTACCAAAACAGGCACAGCATATACTTTCTTTACACCTAATAACATAAAGCAAGCGA
GCGACCTTATCTCTGTGCTTCGGGAAGCTAATCAAGCAATTAATCCCAAGTTGCTTCAGTTGGTCGAAGACAGAGGTTCA
GGTCGTTCCAGGGGTAGAGGAGGCATGAAGGACGATCGTCGTGACAGATACTCTGCAGGCAAAAGGGGTGGATTTAATAC
CTTTAGAGACAGGGAAAACTATGACAGAGGCTACTCTAATCTGCTTAAGAGAGATTTTGGGGCTAAAACTCAGAATGATG
TTTACAGTACTGCAAATTACACCAATGGGAGCTTTGGAAGTAATTTTGTATCTGCTGGCATACAGACCAGTTTTAGGACT
GGTAATCCAACAGGGACTTACCAGAACGATTATGATAGCACTCAGCAATATGGAAGTAATGTTGCAAATATGCACAATGG
TATGAACCAACAGGCATATGCGTATCCTGCTACCGCAGCTGCTGCGCCTATGATTGGCTATCCCGTGCCAACAGGGTATT
CTCAA
ATGTCGAGTTATTCTAGTGACCGAGACCACAGCCGGGATCGAGGGTTTGGTGCACCTCGATTTGGAGGGAGTAGAACAGG
ACCCCTCTCTGGAAAGAAGTTTGGAAATCCTGGGGAGAAACTAGTTAAAAAGAAGTGGAATCTTGATGAGCTGCCCAAAT
TTGAGAAGAATTTTTATCAAGAACACCCTGATTTGGCAAGGCGCACCGCACAAGAGGTTGATACATACAGAAGAAGCAAG
GAAATTACAGTTAGAGGTCACAACTGTCCAAAACCTGTTCTGAAGTTTTATGAAGCAAACTTTCCTGCGAATGTCATGGA
CGTGATTGCAAGGCAGAACTTCACTGAACCCACTGCTATTCAAGCTCAGGGCTGGCCAGTTGCTCTCAGTGGATTGGATA
TGGTTGGAGTAGCTCAGACTGGATCTGGGAAAACACTATCTTATTTGCTGCCTGCCATTGTACACATAATCCACCAGCCA
TTCCTAGAGAGAGGTGATGGGCCTATTTGCTTGGTGCTGGCACCAACTCGAGAACTGGCACAGCAGGTGCAGCAAGTGGC
TGCTGAATATTGTAGAGCTTGTCGCTTGAAGTCTACTTGCATCTATGGTGGTGCTCCCAAAGGACCACAGATTCGTGATT
TGGAAAGAGGCGTGGAAATCTGTATTGCAACACCTGGAAGACTGATTGACTTTTTAGAGTGTGGGAAAACCAATCTGAGA
AGAACAACGTACCTTGTCCTCGATGAAGCTGATAGGATGCTTGATATGGGATTTGAACCCCAGATAAGGAAAATCGTGGA
TCAAATAAGACCTGATAGGCAAACACTAATGTGGAGTGCAACTTGGCCAAAAGAAGTAAGACAGCTTGCTGAAGATTTCC
TGAAAGACTATATTCATATCAATATTGGTGCGCTGGAACTGAGTGCAAACCATAACATTCTCCAGATTGTGGATGTATGT
CATGATGTCGAAAAGGATGAAAAGCTTATTCTTCTGATGGAAGAAATCATGAGTGAGAAGGAGAATAAAACCATTGTTTT
TGTTGAAACCAAAAGAAGATGTGATGAACTTACCAGAAAAATGAGGAGAGATGGGTGGCCTGCCATGGGCATCCATGGTG
ACAAGAGTCAGCAGGAACGTGACTGGGTTCTAAGTGAATTCAAACATGGAAAAGCTTCTATTCTGATTGCTACTGATGTG
GCCTCCAGAGGGCTAGATGTGGAAGATGTGAAATTTGTCATCAATTATGACTACCCTAACTCCTCAGAGGATTATATTCA
TCGAATTGGAAGAACTGCTCGCAGTACCAAAACAGGCACAGCATATACTTTCTTTACACCTAATAACATAAAGCAAGCGA
GCGACCTTATCTCTGTGCTTCGGGAAGCTAATCAAGCAATTAATCCCAAGTTGCTTCAGTTGGTCGAAGACAGAGGTTCA
GGTCGTTCCAGGGGTAGAGGAGGCATGAAGGACGATCGTCGTGACAGATACTCTGCAGGCAAAAGGGGTGGATTTAATAC
CTTTAGAGACAGGGAAAACTATGACAGAGGCTACTCTAATCTGCTTAAGAGAGATTTTGGGGCTAAAACTCAGAATGATG
TTTACAGTACTGCAAATTACACCAATGGGAGCTTTGGAAGTAATTTTGTATCTGCTGGCATACAGACCAGTTTTAGGACT
GGTAATCCAACAGGGACTTACCAGAACGATTATGATAGCACTCAGCAATATGGAAGTAATGTTGCAAATATGCACAATGG
TATGAACCAACAGGCATATGCGTATCCTGCTACCGCAGCTGCTGCGCCTATGATTGGCTATCCCGTGCCAACAGGGTATT
CTCAA
ORF - retro_mmus_788 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 98.05 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MSSYSSDRDRGRDRGFGAPRFGGSRTGPLSGKKFGNPGEKLVKKKWNLDELPKFEKNFYQEHPDLARRTA |
| MSSYSSDRD..RDRGFGAPRFGGSRTGPLSGKKFGNPGEKLVKKKWNLDELPKFEKNFYQEHPDLARRTA | |
| Retrocopy | MSSYSSDRDHSRDRGFGAPRFGGSRTGPLSGKKFGNPGEKLVKKKWNLDELPKFEKNFYQEHPDLARRTA |
| Parental | QEVDTYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARQNFTEPTAIQAQGWPVALSGLDMVGVAQT |
| QEVDTYRRSKEITVRGHNCPKPVL.FYEANFPANVMDVIARQNFTEPTAIQAQGWPVALSGLDMVGVAQT | |
| Retrocopy | QEVDTYRRSKEITVRGHNCPKPVLKFYEANFPANVMDVIARQNFTEPTAIQAQGWPVALSGLDMVGVAQT |
| Parental | GSGKTLSYLLPAIVHINHQPFLERGDGPICLVLAPTRELAQQVQQVAAEYCRACRLKSTCIYGGAPKGPQ |
| GSGKTLSYLLPAIVHI.HQPFLERGDGPICLVLAPTRELAQQVQQVAAEYCRACRLKSTCIYGGAPKGPQ | |
| Retrocopy | GSGKTLSYLLPAIVHIIHQPFLERGDGPICLVLAPTRELAQQVQQVAAEYCRACRLKSTCIYGGAPKGPQ |
| Parental | IRDLERGVEICIATPGRLIDFLECGKTNLRRTTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSA |
| IRDLERGVEICIATPGRLIDFLECGKTNLRRTTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSA | |
| Retrocopy | IRDLERGVEICIATPGRLIDFLECGKTNLRRTTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSA |
| Parental | TWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEKDEKLIRLMEEIMSEKENKTIVFVET |
| TWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEKDEKLI.LMEEIMSEKENKTIVFVET | |
| Retrocopy | TWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEKDEKLILLMEEIMSEKENKTIVFVET |
| Parental | KRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVLNEFKHGKAPILIATDVASRGLDVEDVKFVINYDYPN |
| KRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVL.EFKHGKA.ILIATDVASRGLDVEDVKFVINYDYPN | |
| Retrocopy | KRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVLSEFKHGKASILIATDVASRGLDVEDVKFVINYDYPN |
| Parental | SSEDYIHRIGRTARSTKTGTAYTFFTPNNIKQVSDLISVLREANQAINPKLLQLVEDRGSGRSRGRGGMK |
| SSEDYIHRIGRTARSTKTGTAYTFFTPNNIKQ.SDLISVLREANQAINPKLLQLVEDRGSGRSRGRGGMK | |
| Retrocopy | SSEDYIHRIGRTARSTKTGTAYTFFTPNNIKQASDLISVLREANQAINPKLLQLVEDRGSGRSRGRGGMK |
| Parental | DDRRDRYSAGKRGGFNTFRDRENYDRGYSNLLKRDFGAKTQNGVYSAANYTNGSFGSNFVSAGIQTSFRT |
| DDRRDRYSAGKRGGFNTFRDRENYDRGYSNLLKRDFGAKTQN.VYS.ANYTNGSFGSNFVSAGIQTSFRT | |
| Retrocopy | DDRRDRYSAGKRGGFNTFRDRENYDRGYSNLLKRDFGAKTQNDVYSTANYTNGSFGSNFVSAGIQTSFRT |
| Parental | GNPTGTYQNGYDSTQQYGSNVANMHNGMNQQAYAYPATAAAAPMIGYPMPTGYSQ |
| GNPTGTYQN.YDSTQQYGSNVANMHNGMNQQAYAYPATAAAAPMIGYP.PTGYSQ | |
| Retrocopy | GNPTGTYQNDYDSTQQYGSNVANMHNGMNQQAYAYPATAAAAPMIGYPVPTGYSQ |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 1 .50 RPM | 645 .89 RPM |
| SRP007412_cerebellum | 1 .52 RPM | 930 .74 RPM |
| SRP007412_heart | 1 .12 RPM | 347 .91 RPM |
| SRP007412_kidney | 1 .81 RPM | 579 .55 RPM |
| SRP007412_liver | 0 .66 RPM | 372 .57 RPM |
| SRP007412_testis | 1 .78 RPM | 661 .48 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_788 was not detected
No EST(s) were mapped for retro_mmus_788 retrocopy.
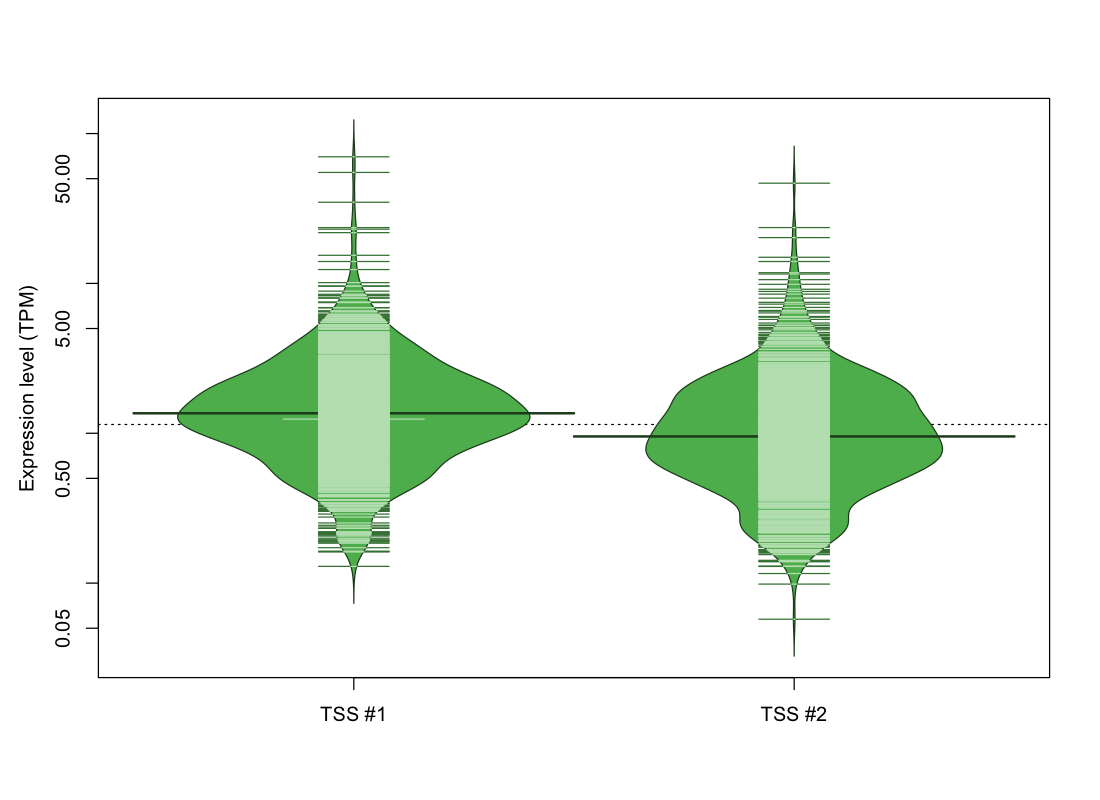
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_12942 | 149 libraries | 311 libraries | 564 libraries | 38 libraries | 10 libraries |
| TSS #2 | TSS_12943 | 207 libraries | 455 libraries | 381 libraries | 21 libraries | 8 libraries |
The graphical summary, for retro_mmus_788 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_788 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_788 has 0 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 12 parental genes, and 19 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Echinops telfairi | ENSETEG00000019950 | 1 retrocopy | |
| Felis catus | ENSFCAG00000003512 | 1 retrocopy | |
| Myotis lucifugus | ENSMLUG00000009496 | 4 retrocopies | |
| Monodelphis domestica | ENSMODG00000003743 | 4 retrocopies | |
| Mus musculus | ENSMUSG00000020075 | 2 retrocopies | |
| Mus musculus | ENSMUSG00000020076 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000020719 | 1 retrocopy |
retro_mmus_788 ,
|
| Pteropus vampyrus | ENSPVAG00000001932 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000030680 | 1 retrocopy | |
| Tupaia belangeri | ENSTBEG00000009490 | 1 retrocopy | |
| Tarsius syrichta | ENSTSYG00000012149 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000008793 | 1 retrocopy |