RetrogeneDB ID: | retro_hsap_36 | ||
Retrocopylocation | Organism: | Human (Homo sapiens) | |
| Coordinates: | 14:50360454..50360982(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSG00000165527 | |
| Aliases: | None | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | ARF3 | ||
| Ensembl ID: | ENSG00000134287 | ||
| Aliases: | None | ||
| Description: | ADP-ribosylation factor 3 [Source:HGNC Symbol;Acc:654] |

Retrocopy-Parental alignment summary:
>retro_hsap_36
ATGGGGAAGGTGCTATCCAAAATCTTCGGGAACAAGGAAATGCGGATCCTCATGTTGGGCCTGGACGCGGCCGGCAAGAC
AACAATCCTGTACAAGTTGAAGCTGGGCCAGTCGGTGACCACCATTCCCACTGTGGGTTTCAACGTGGAGACGGTGACTT
ACAAAAATGTCAAGTTCAACGTATGGGATGTGGGCGGCCAGGACAAGATCCGGCCGCTCTGGCGGCATTACTACACTGGG
ACCCAAGGTCTCATCTTCGTAGTGGACTGCGCCGACCGCGACCGCATCGATGAGGCTCGCCAGGAGCTGCACCGCATTAT
CAATGACCGGGAGATGAGGGACGCCATAATCCTCATCTTCGCCAACAAGCAGGACCTGCCCGATGCCATGAAACCCCACG
AGATCCAGGAGAAACTGGGCCTGACCCGGATTCGGGACAGGAACTGGTATGTGCAGCCCTCCTGTGCCACCTCAGGGGAC
GGACTCTATGAGGGGCTCACATGGTTAACCTCTAACTACAAATCTTAA
ATGGGGAAGGTGCTATCCAAAATCTTCGGGAACAAGGAAATGCGGATCCTCATGTTGGGCCTGGACGCGGCCGGCAAGAC
AACAATCCTGTACAAGTTGAAGCTGGGCCAGTCGGTGACCACCATTCCCACTGTGGGTTTCAACGTGGAGACGGTGACTT
ACAAAAATGTCAAGTTCAACGTATGGGATGTGGGCGGCCAGGACAAGATCCGGCCGCTCTGGCGGCATTACTACACTGGG
ACCCAAGGTCTCATCTTCGTAGTGGACTGCGCCGACCGCGACCGCATCGATGAGGCTCGCCAGGAGCTGCACCGCATTAT
CAATGACCGGGAGATGAGGGACGCCATAATCCTCATCTTCGCCAACAAGCAGGACCTGCCCGATGCCATGAAACCCCACG
AGATCCAGGAGAAACTGGGCCTGACCCGGATTCGGGACAGGAACTGGTATGTGCAGCCCTCCTGTGCCACCTCAGGGGAC
GGACTCTATGAGGGGCTCACATGGTTAACCTCTAACTACAAATCTTAA
ORF - retro_hsap_36 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 69.54 % |
| Parental protein coverage: | 96.13 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | GNLLKSLIGKKEMRILMVGLDAAGKTTILYKLKLGEIVTTIPTIGFNVETVEYKNISFTVWDVGGQDKIR |
| G..L....G.KEMRILM.GLDAAGKTTILYKLKLG..VTTIPT.GFNVETV.YKN..F.VWDVGGQDKIR | |
| Retrocopy | GKVLSKIFGNKEMRILMLGLDAAGKTTILYKLKLGQSVTTIPTVGFNVETVTYKNVKFNVWDVGGQDKIR |
| Parental | PLWRHYFQNTQGLIFVVDSNDRERVNEAREELMRMLAEDELRDAVLLVFANKQDLPNAMNAAEITDKLGL |
| PLWRHY...TQGLIFVVD..DR.R..EAR.EL.R.....E.RDA..L.FANKQDLP.AM...EI..KLGL | |
| Retrocopy | PLWRHYYTGTQGLIFVVDCADRDRIDEARQELHRIINDREMRDAIILIFANKQDLPDAMKPHEIQEKLGL |
| Parental | HSLRHRNWYIQATCATSGDGLYEGLDWLANQLKN |
| ...R.RNWY.Q..CATSGDGLYEGL.WL....K. | |
| Retrocopy | TRIRDRNWYVQPSCATSGDGLYEGLTWLTSNYKS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| bodymap2_adipose | 29 .74 RPM | 222 .28 RPM |
| bodymap2_adrenal | 36 .74 RPM | 202 .28 RPM |
| bodymap2_brain | 9 .21 RPM | 477 .85 RPM |
| bodymap2_breast | 21 .67 RPM | 155 .62 RPM |
| bodymap2_colon | 16 .53 RPM | 182 .88 RPM |
| bodymap2_heart | 8 .92 RPM | 43 .75 RPM |
| bodymap2_kidney | 23 .35 RPM | 192 .83 RPM |
| bodymap2_liver | 13 .03 RPM | 55 .17 RPM |
| bodymap2_lung | 32 .05 RPM | 215 .07 RPM |
| bodymap2_lymph_node | 35 .18 RPM | 138 .22 RPM |
| bodymap2_ovary | 17 .83 RPM | 206 .47 RPM |
| bodymap2_prostate | 25 .17 RPM | 152 .09 RPM |
| bodymap2_skeletal_muscle | 21 .01 RPM | 17 .62 RPM |
| bodymap2_testis | 21 .00 RPM | 221 .73 RPM |
| bodymap2_thyroid | 16 .95 RPM | 265 .03 RPM |
| bodymap2_white_blood_cells | 64 .67 RPM | 309 .73 RPM |
RNA Polymerase II actvity may be related with retro_hsap_36 in 52 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001VKU | POLR2A | 14:50359215..50360263 |
| ENCFF002CFW | POLR2A | 14:50359310..50360442 |
| ENCFF002CFX | POLR2A | 14:50359317..50360285 |
| ENCFF002CGN | POLR2A | 14:50359419..50360032 |
| ENCFF002CHO | POLR2A | 14:50358989..50361061 |
| ENCFF002CIH | POLR2A | 14:50359697..50360022 |
| ENCFF002CIH | POLR2A | 14:50359459..50359724 |
| ENCFF002CIO | POLR2A | 14:50359397..50360427 |
| ENCFF002CJE | POLR2A | 14:50359322..50360353 |
| ENCFF002CJZ | POLR2A | 14:50359258..50360341 |
| ENCFF002CKX | POLR2A | 14:50359403..50360032 |
| ENCFF002CLM | POLR2A | 14:50359797..50359932 |
| ENCFF002CMI | POLR2A | 14:50359368..50360492 |
| ENCFF002COJ | POLR2A | 14:50359604..50360040 |
| ENCFF002CPG | POLR2A | 14:50359455..50359748 |
| ENCFF002CPG | POLR2A | 14:50359686..50360004 |
| ENCFF002CPH | POLR2A | 14:50359425..50359815 |
| ENCFF002CPH | POLR2A | 14:50359645..50360035 |
| ENCFF002CQA | POLR2A | 14:50359446..50359774 |
| ENCFF002CQA | POLR2A | 14:50359765..50360008 |
| ENCFF002CQC | POLR2A | 14:50359419..50360032 |
| ENCFF002CQE | POLR2A | 14:50359394..50360036 |
| ENCFF002CQG | POLR2A | 14:50359467..50360013 |
| ENCFF002CQI | POLR2A | 14:50359415..50360025 |
| ENCFF002CQK | POLR2A | 14:50359384..50360051 |
| ENCFF002CQM | POLR2A | 14:50359424..50360038 |
| ENCFF002CQO | POLR2A | 14:50359360..50360388 |
| ENCFF002CRK | POLR2A | 14:50359352..50359876 |
| ENCFF002CRK | POLR2A | 14:50359560..50360084 |
| ENCFF002CRO | POLR2A | 14:50359390..50359810 |
| ENCFF002CRO | POLR2A | 14:50359742..50359948 |
| ENCFF002CSY | POLR2A | 14:50358981..50360515 |
| ENCFF002CUP | POLR2A | 14:50359667..50359947 |
| ENCFF002CUQ | POLR2A | 14:50359404..50360036 |
| ENCFF002CVF | POLR2A | 14:50359747..50359902 |
| ENCFF002CVF | POLR2A | 14:50359409..50359825 |
| ENCFF002CVJ | POLR2A | 14:50359400..50360354 |
| ENCFF002CXM | POLR2A | 14:50359584..50360074 |
| ENCFF002CXN | POLR2A | 14:50359334..50359830 |
| ENCFF002CXN | POLR2A | 14:50359610..50360106 |
| ENCFF002CXO | POLR2A | 14:50359733..50359910 |
| ENCFF002CXO | POLR2A | 14:50359522..50359672 |
| ENCFF002CXP | POLR2A | 14:50359696..50359982 |
| ENCFF002CXR | POLR2A | 14:50359439..50359771 |
| ENCFF002CXR | POLR2A | 14:50359670..50359995 |
| ENCFF002CZC | POLR2A | 14:50359373..50360064 |
| ENCFF002CZD | POLR2A | 14:50359393..50360060 |
| ENCFF002CZQ | POLR2A | 14:50359674..50360004 |
| ENCFF002CZQ | POLR2A | 14:50359442..50359763 |
| ENCFF002CZW | POLR2A | 14:50359261..50360273 |
| ENCFF002CZY | POLR2A | 14:50359378..50360049 |
| ENCFF002DAE | POLR2A | 14:50359555..50359633 |
| ENCFF002DAE | POLR2A | 14:50359782..50359881 |
| ENCFF002DAH | POLR2A | 14:50359691..50359975 |
| ENCFF002DAK | POLR2A | 14:50359342..50359842 |
| ENCFF002DAK | POLR2A | 14:50359543..50360043 |
| ENCFF002DAS | POLR2A | 14:50359364..50359840 |
| ENCFF002DAS | POLR2A | 14:50359593..50360069 |
| ENCFF002DAV | POLR2A | 14:50359339..50359883 |
| ENCFF002DAV | POLR2A | 14:50359772..50359919 |
| ENCFF002DAY | POLR2A | 14:50359278..50359914 |
| ENCFF002DAY | POLR2A | 14:50359520..50360156 |
| ENCFF002DBB | POLR2A | 14:50359503..50359703 |
| ENCFF002DBB | POLR2A | 14:50359774..50359974 |
| ENCFF002DBE | POLR2A | 14:50359693..50359967 |
| ENCFF002DBO | POLR2A | 14:50359432..50360035 |
| ENCFF002DBP | POLR2A | 14:50359464..50359746 |
| ENCFF002DBP | POLR2A | 14:50359696..50359983 |
| ENCFF002DBQ | POLR2A | 14:50359515..50359713 |
| ENCFF002DBQ | POLR2A | 14:50359758..50359897 |
| ENCFF002DBT | POLR2A | 14:50359585..50359677 |
| ENCFF002DBT | POLR2A | 14:50359804..50359897 |
29 EST(s) were mapped to retro_hsap_36 retrocopy
| EST ID | Start | End | Identity | Match | Mis-match | Score |
|---|---|---|---|---|---|---|
| AI940133 | 50360534 | 50360708 | 95.3 | 164 | 4 | 154 |
| AI940136 | 50360551 | 50360660 | 98.1 | 105 | 0 | 100 |
| AW751295 | 50360566 | 50360693 | 97.6 | 122 | 3 | 117 |
| AW751296 | 50360551 | 50360666 | 98.2 | 107 | 1 | 102 |
| BQ315629 | 50360533 | 50360696 | 95.1 | 159 | 3 | 153 |
| BQ315631 | 50360525 | 50360735 | 99.6 | 203 | 1 | 196 |
| BQ315632 | 50360556 | 50360725 | 98.3 | 164 | 3 | 159 |
| BQ315633 | 50360542 | 50360724 | 99.5 | 175 | 0 | 169 |
| BQ315634 | 50360562 | 50360720 | 99.4 | 157 | 1 | 156 |
| BQ315635 | 50360576 | 50360724 | 99.4 | 147 | 1 | 146 |
| BQ315636 | 50360539 | 50360735 | 99 | 188 | 2 | 181 |
| BQ315637 | 50360536 | 50360724 | 99 | 181 | 1 | 175 |
| BQ315638 | 50360573 | 50360726 | 99.4 | 150 | 0 | 147 |
| BQ315639 | 50360543 | 50360724 | 98.3 | 171 | 2 | 164 |
| BQ315640 | 50360547 | 50360714 | 100 | 163 | 0 | 159 |
| BQ315641 | 50360547 | 50360724 | 99.4 | 164 | 0 | 157 |
| BQ315642 | 50360527 | 50360724 | 99.5 | 190 | 1 | 184 |
| BQ315643 | 50360517 | 50360715 | 97.9 | 183 | 3 | 174 |
| BQ315644 | 50360565 | 50360724 | 99.4 | 158 | 1 | 157 |
| BQ315645 | 50360532 | 50360724 | 99.5 | 186 | 1 | 182 |
| BQ315646 | 50360542 | 50360724 | 98.9 | 173 | 1 | 167 |
| BQ315647 | 50360575 | 50360724 | 100 | 148 | 0 | 147 |
| BQ315648 | 50360551 | 50360728 | 99.5 | 175 | 1 | 173 |
| BQ315707 | 50360529 | 50360724 | 100 | 195 | 0 | 195 |
| BQ315714 | 50360529 | 50360724 | 100 | 195 | 0 | 195 |
| BQ315722 | 50360519 | 50360693 | 98.3 | 169 | 3 | 164 |
| BQ315746 | 50360584 | 50360735 | 97.2 | 138 | 2 | 132 |
| BQ315747 | 50360581 | 50360717 | 97 | 130 | 1 | 124 |
| EC481026 | 50360489 | 50360592 | 100 | 103 | 0 | 103 |
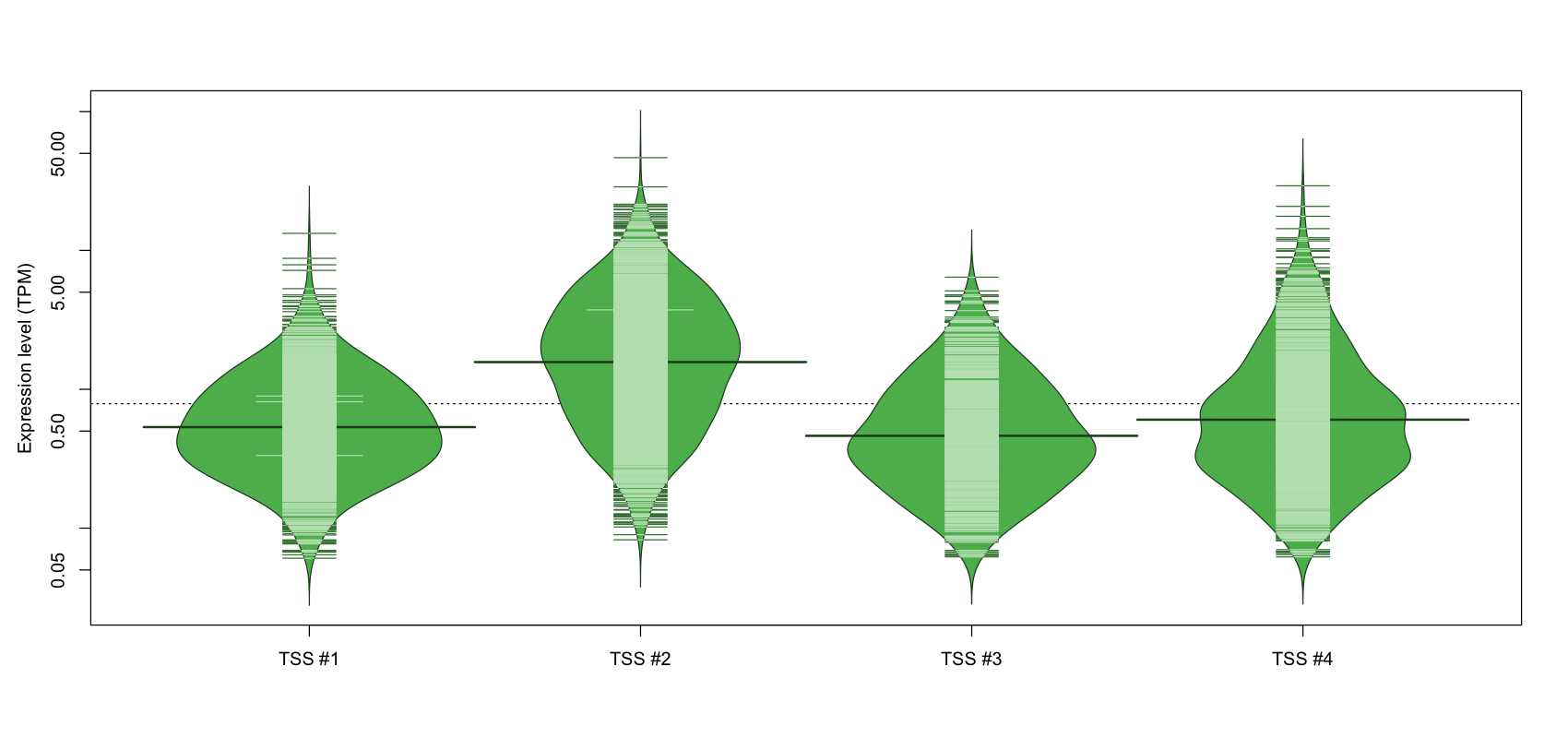
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_38365 | 743 libraries | 835 libraries | 246 libraries | 4 libraries | 1 library |
| TSS #2 | TSS_38366 | 342 libraries | 521 libraries | 735 libraries | 180 libraries | 51 libraries |
| TSS #3 | TSS_38369 | 1141 libraries | 544 libraries | 142 libraries | 2 libraries | 0 libraries |
| TSS #4 | TSS_38370 | 919 libraries | 643 libraries | 229 libraries | 30 libraries | 8 libraries |
The graphical summary, for retro_hsap_36 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1829 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_hsap_36 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_hsap_36 has 12 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 20 parental genes, and 31 retrocopies.Expression level across human populations :
| Library | Retrogene expression |
|---|---|
| CEU_NA11831 | 56 .09 RPM |
| CEU_NA11843 | 52 .38 RPM |
| CEU_NA11930 | 46 .84 RPM |
| CEU_NA12004 | 60 .02 RPM |
| CEU_NA12400 | 50 .72 RPM |
| CEU_NA12751 | 58 .16 RPM |
| CEU_NA12760 | 61 .57 RPM |
| CEU_NA12827 | 63 .77 RPM |
| CEU_NA12872 | 60 .37 RPM |
| CEU_NA12873 | 65 .69 RPM |
| FIN_HG00183 | 46 .87 RPM |
| FIN_HG00277 | 51 .87 RPM |
| FIN_HG00315 | 63 .29 RPM |
| FIN_HG00321 | 48 .09 RPM |
| FIN_HG00328 | 65 .67 RPM |
| FIN_HG00338 | 58 .80 RPM |
| FIN_HG00349 | 58 .47 RPM |
| FIN_HG00375 | 56 .83 RPM |
| FIN_HG00377 | 67 .17 RPM |
| FIN_HG00378 | 66 .51 RPM |
| GBR_HG00099 | 69 .48 RPM |
| GBR_HG00111 | 55 .17 RPM |
| GBR_HG00114 | 71 .05 RPM |
| GBR_HG00119 | 52 .71 RPM |
| GBR_HG00131 | 46 .71 RPM |
| GBR_HG00133 | 62 .25 RPM |
| GBR_HG00134 | 66 .17 RPM |
| GBR_HG00137 | 60 .42 RPM |
| GBR_HG00142 | 48 .07 RPM |
| GBR_HG00143 | 66 .75 RPM |
| TSI_NA20512 | 75 .17 RPM |
| TSI_NA20513 | 57 .19 RPM |
| TSI_NA20518 | 62 .53 RPM |
| TSI_NA20532 | 56 .87 RPM |
| TSI_NA20538 | 61 .87 RPM |
| TSI_NA20756 | 62 .97 RPM |
| TSI_NA20765 | 57 .62 RPM |
| TSI_NA20771 | 62 .81 RPM |
| TSI_NA20786 | 72 .42 RPM |
| TSI_NA20798 | 77 .12 RPM |
| YRI_NA18870 | 74 .03 RPM |
| YRI_NA18907 | 70 .33 RPM |
| YRI_NA18916 | 68 .20 RPM |
| YRI_NA19093 | 92 .25 RPM |
| YRI_NA19099 | 75 .79 RPM |
| YRI_NA19114 | 65 .14 RPM |
| YRI_NA19118 | 70 .98 RPM |
| YRI_NA19213 | 65 .39 RPM |
| YRI_NA19214 | 90 .02 RPM |
| YRI_NA19223 | 71 .09 RPM |