RetrogeneDB ID: | retro_mmus_147 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 12:69372793..69373321(+) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000044147 | |
| Aliases: | Arf6, AI788669, AW496366 | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Arf3 | ||
| Ensembl ID: | ENSMUSG00000051853 | ||
| Aliases: | Arf3, 5430400P17Rik, AI854770 | ||
| Description: | ADP-ribosylation factor 3 [Source:MGI Symbol;Acc:MGI:99432] |

Retrocopy-Parental alignment summary:
>retro_mmus_147
ATGGGGAAGGTGCTATCCAAGATCTTCGGGAACAAGGAAATGCGGATCCTCATGCTGGGCCTGGACGCAGCCGGCAAGAC
AACGATCCTGTACAAGTTGAAGCTGGGCCAATCGGTGACCACCATCCCCACGGTGGGCTTCAACGTGGAGACGGTGACTT
ACAAAAACGTCAAGTTCAACGTGTGGGATGTGGGCGGCCAGGACAAGATCCGGCCGCTCTGGCGGCATTACTACACCGGG
ACCCAGGGTCTGATCTTCGTGGTAGACTGCGCCGACCGCGACCGCATCGACGAGGCCCGCCAGGAGCTGCACCGCATTAT
CAATGACCGGGAGATGAGGGACGCCATCATCCTCATCTTCGCCAACAAGCAGGACCTGCCCGATGCCATGAAACCCCATG
AGATCCAGGAGAAACTGGGCCTGACCCGGATTCGGGACAGGAACTGGTATGTGCAGCCCTCCTGTGCCACCTCCGGGGAC
GGACTCTATGAGGGGCTCACATGGTTAACCTCTAACTACAAATCCTAA
ATGGGGAAGGTGCTATCCAAGATCTTCGGGAACAAGGAAATGCGGATCCTCATGCTGGGCCTGGACGCAGCCGGCAAGAC
AACGATCCTGTACAAGTTGAAGCTGGGCCAATCGGTGACCACCATCCCCACGGTGGGCTTCAACGTGGAGACGGTGACTT
ACAAAAACGTCAAGTTCAACGTGTGGGATGTGGGCGGCCAGGACAAGATCCGGCCGCTCTGGCGGCATTACTACACCGGG
ACCCAGGGTCTGATCTTCGTGGTAGACTGCGCCGACCGCGACCGCATCGACGAGGCCCGCCAGGAGCTGCACCGCATTAT
CAATGACCGGGAGATGAGGGACGCCATCATCCTCATCTTCGCCAACAAGCAGGACCTGCCCGATGCCATGAAACCCCATG
AGATCCAGGAGAAACTGGGCCTGACCCGGATTCGGGACAGGAACTGGTATGTGCAGCCCTCCTGTGCCACCTCCGGGGAC
GGACTCTATGAGGGGCTCACATGGTTAACCTCTAACTACAAATCCTAA
ORF - retro_mmus_147 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 69.54 % |
| Parental protein coverage: | 96.13 % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | GNLLKSLIGKKEMRILMVGLDAAGKTTILYKLKLGEIVTTIPTIGFNVETVEYKNISFTVWDVGGQDKIR |
| G..L....G.KEMRILM.GLDAAGKTTILYKLKLG..VTTIPT.GFNVETV.YKN..F.VWDVGGQDKIR | |
| Retrocopy | GKVLSKIFGNKEMRILMLGLDAAGKTTILYKLKLGQSVTTIPTVGFNVETVTYKNVKFNVWDVGGQDKIR |
| Parental | PLWRHYFQNTQGLIFVVDSNDRERVNEAREELMRMLAEDELRDAVLLVFANKQDLPNAMNAAEITDKLGL |
| PLWRHY...TQGLIFVVD..DR.R..EAR.EL.R.....E.RDA..L.FANKQDLP.AM...EI..KLGL | |
| Retrocopy | PLWRHYYTGTQGLIFVVDCADRDRIDEARQELHRIINDREMRDAIILIFANKQDLPDAMKPHEIQEKLGL |
| Parental | HSLRHRNWYIQATCATSGDGLYEGLDWLANQLKN |
| ...R.RNWY.Q..CATSGDGLYEGL.WL....K. | |
| Retrocopy | TRIRDRNWYVQPSCATSGDGLYEGLTWLTSNYKS |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 9 .70 RPM | 1302 .01 RPM |
| SRP007412_cerebellum | 9 .86 RPM | 497 .61 RPM |
| SRP007412_heart | 11 .02 RPM | 58 .90 RPM |
| SRP007412_kidney | 15 .15 RPM | 244 .53 RPM |
| SRP007412_liver | 15 .40 RPM | 102 .00 RPM |
| SRP007412_testis | 9 .88 RPM | 157 .80 RPM |
RNA Polymerase II actvity may be related with retro_mmus_147 in 4 libraries
| ENCODE library ID | Target | ChIP-Seq Peak coordinates |
|---|---|---|
| ENCFF001XVA | POLR2A | 12:69371644..69373409 |
| ENCFF001YIJ | POLR2A | 12:69370873..69373493 |
| ENCFF001YJZ | POLR2A | 12:69370910..69372728 |
| ENCFF001YKA | POLR2A | 12:69370883..69373491 |
No EST(s) were mapped for retro_mmus_147 retrocopy.
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_24002 | 86 libraries | 185 libraries | 589 libraries | 177 libraries | 35 libraries |
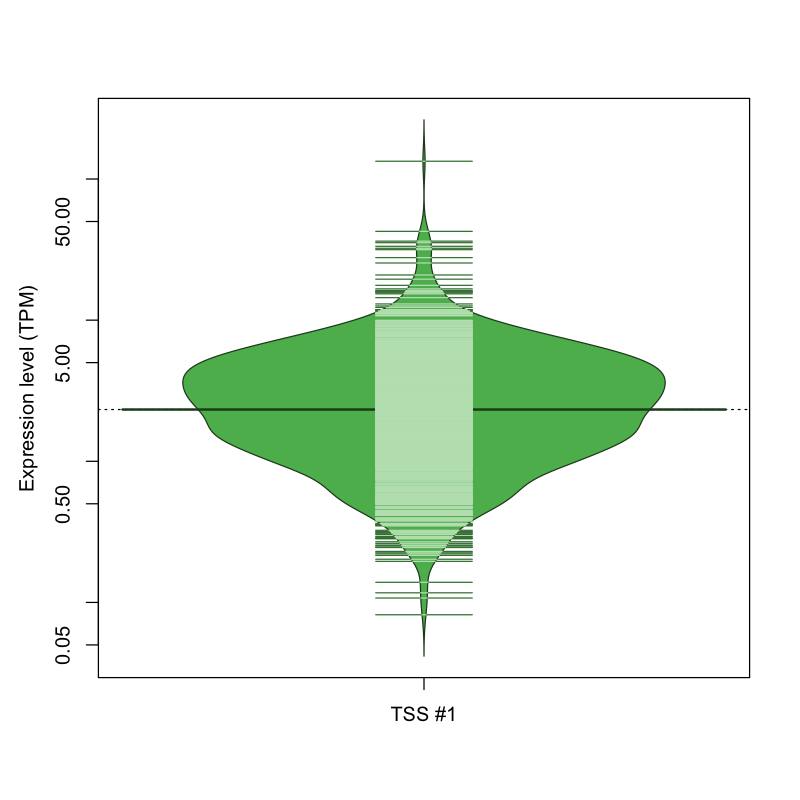
The graphical summary, for retro_mmus_147 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_147 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_147 has 12 orthologous retrocopies within eutheria group .Parental genes homology:
Parental genes homology involve 19 parental genes, and 27 retrocopies.| Species | Parental gene accession | Retrocopies number | |
|---|---|---|---|
| Anolis carolinensis | ENSACAG00000008240 | 1 retrocopy | |
| Cavia porcellus | ENSCPOG00000025074 | 1 retrocopy | |
| Ciona savignyi | ENSCSAVG00000007011 | 1 retrocopy | |
| Homo sapiens | ENSG00000134287 | 1 retrocopy | |
| Latimeria chalumnae | ENSLACG00000019009 | 1 retrocopy | |
| Loxodonta africana | ENSLAFG00000004207 | 1 retrocopy | |
| Macropus eugenii | ENSMEUG00000005072 | 7 retrocopies | |
| Microcebus murinus | ENSMICG00000017545 | 1 retrocopy | |
| Monodelphis domestica | ENSMODG00000013804 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000048076 | 1 retrocopy | |
| Mus musculus | ENSMUSG00000051853 | 1 retrocopy |
retro_mmus_147 ,
|
| Mus musculus | ENSMUSG00000062421 | 1 retrocopy | |
| Nomascus leucogenys | ENSNLEG00000017859 | 2 retrocopies | |
| Petromyzon marinus | ENSPMAG00000000531 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000033155 | 1 retrocopy | |
| Rattus norvegicus | ENSRNOG00000048089 | 1 retrocopy | |
| Sarcophilus harrisii | ENSSHAG00000003635 | 2 retrocopies | |
| Tetraodon nigroviridis | ENSTNIG00000011685 | 1 retrocopy | |
| Tursiops truncatus | ENSTTRG00000004698 | 1 retrocopy |