RetrogeneDB ID: | retro_mmus_258 | ||
Retrocopylocation | Organism: | Mouse (Mus musculus) | |
| Coordinates: | 10:99757800..99759201(-) | ||
| Located in intron of: | None | ||
Retrocopyinformation | Ensembl ID: | ENSMUSG00000046934 | |
| Aliases: | Csl, 1700007H16Rik | ||
| Status: | KNOWN_PROTEIN_CODING | ||
Parental geneinformation | Parental gene summary: | ||
| Parental gene symbol: | Cs | ||
| Ensembl ID: | ENSMUSG00000005683 | ||
| Aliases: | Cs, 2610511A05Rik, 9030605P22Rik, BB234005, Cis, ahl4 | ||
| Description: | citrate synthase [Source:MGI Symbol;Acc:MGI:88529] |

Retrocopy-Parental alignment summary:
>retro_mmus_258
ATGGCTCTACTTACTGCGGCAGCCTGGTTCTTGGGAACCAAAAACCCACCCTGCCTTGTCCTTGCCGCCCGGCATGCCAG
TGCTTCTTCCACGAACTTGAAAGACGTGCTGAGGAATCTGATACCCAAGGAGCAAGCCAGAATTAAGACCTTCAGAAAGA
AACACGGGAAGACAGTTGTGGGCCAGATCACTGTGGACATGATGTACGGTGGCATGAGAGGCATGAAGGGACTTGTGTAT
GAGACATCGGTTCTTGATCCTGATGAGGGCATCCGTTTCCGAGGCTACAGTATCCCTGAGTGCCAGAAACTGCTGCCTAA
GGCTAAGGGTGGGAAAGAACCCCTGCCTGAGGGCTTATTTTGGCTGCTGGTAACTGGACAGATGCCCACAGAGGAACAGG
TATCTTGGCTCTCACAAGAATGGGTAAAAAGGGCAGCTCTACCTTCTCATGTCGTCACCATGCTGGACAATTTTCCGACC
AAATTGCACCCCATGTCTCAGCTCAGTGCAGCCATCACAGTCCTCAACAATGAAAGCAACTTTGCCCGGGCATATGCGCA
GGGAATGAACCGAACTAAGTACTGGGAGCTCACCTATGAAGACTGTATGGACCTGCTTGCCAAGCTTCCGTGTGTTGCAG
CAAAGATCTACCGGAATCTGTACCGGGAAGATAGGAATATTGAGGCCATTGACTCTAAGCTGGACTGGTCCCACAATTTT
ACCAACATGTTAGGCTACACGGACCCTCAGTTCACCGAGCTCATGCGTTTGTACCTCACCATCCACAGTGACCATGAGGG
TGGTAATGTAAGTGCCCACACCAGCCATTTGGTGGGCAGTGCCCTTTCTGATCCTTACCTGTCCTTTGCAGCAGCCTTGA
ATGGGCTGGCGGGGCCTCTCCATGGGCTAGCAAATCAGGAGGTGCTTGTCTGGCTGACACAGCTACAGAAGGAAGTTGGC
GAAGATGCATCAGATGAGAAGTTAAAAAACTACATCTGGAACACACTCAACTCAGGACGGGTTGTCCCAGGCTATGGCCA
TGCAGTACTGAGGAAGACTGACCCTCGCTATTCCTGTCAGCGAGAGTTTGCTCTGAAACATCTGCCTAAGGATCCCATGT
TCAAGCTGGTTGGTCAGCTGTACAAGATTGTACCCGATATCCTCTTAGAGCAAGGGAAGGCTAAGAACCCTTGGCCCAAC
GTAGACGCTCACAGTGGGGTGCTGCTCCAGTACTACGGCATGAGGGAGATGAATTACTACACAGTCCTGTTCGGAGTGTC
GCGGGCACTGGGCGTTCTGTCCCAGCTCATCTGGAGCAGAGCCCTAGGCTTCCCTCTGGAAAGGCCCAAGTCCATGAGCA
CGGATGCTCTGATGAAGTTTGTGAACTCTGAGTCAGGATAA
ATGGCTCTACTTACTGCGGCAGCCTGGTTCTTGGGAACCAAAAACCCACCCTGCCTTGTCCTTGCCGCCCGGCATGCCAG
TGCTTCTTCCACGAACTTGAAAGACGTGCTGAGGAATCTGATACCCAAGGAGCAAGCCAGAATTAAGACCTTCAGAAAGA
AACACGGGAAGACAGTTGTGGGCCAGATCACTGTGGACATGATGTACGGTGGCATGAGAGGCATGAAGGGACTTGTGTAT
GAGACATCGGTTCTTGATCCTGATGAGGGCATCCGTTTCCGAGGCTACAGTATCCCTGAGTGCCAGAAACTGCTGCCTAA
GGCTAAGGGTGGGAAAGAACCCCTGCCTGAGGGCTTATTTTGGCTGCTGGTAACTGGACAGATGCCCACAGAGGAACAGG
TATCTTGGCTCTCACAAGAATGGGTAAAAAGGGCAGCTCTACCTTCTCATGTCGTCACCATGCTGGACAATTTTCCGACC
AAATTGCACCCCATGTCTCAGCTCAGTGCAGCCATCACAGTCCTCAACAATGAAAGCAACTTTGCCCGGGCATATGCGCA
GGGAATGAACCGAACTAAGTACTGGGAGCTCACCTATGAAGACTGTATGGACCTGCTTGCCAAGCTTCCGTGTGTTGCAG
CAAAGATCTACCGGAATCTGTACCGGGAAGATAGGAATATTGAGGCCATTGACTCTAAGCTGGACTGGTCCCACAATTTT
ACCAACATGTTAGGCTACACGGACCCTCAGTTCACCGAGCTCATGCGTTTGTACCTCACCATCCACAGTGACCATGAGGG
TGGTAATGTAAGTGCCCACACCAGCCATTTGGTGGGCAGTGCCCTTTCTGATCCTTACCTGTCCTTTGCAGCAGCCTTGA
ATGGGCTGGCGGGGCCTCTCCATGGGCTAGCAAATCAGGAGGTGCTTGTCTGGCTGACACAGCTACAGAAGGAAGTTGGC
GAAGATGCATCAGATGAGAAGTTAAAAAACTACATCTGGAACACACTCAACTCAGGACGGGTTGTCCCAGGCTATGGCCA
TGCAGTACTGAGGAAGACTGACCCTCGCTATTCCTGTCAGCGAGAGTTTGCTCTGAAACATCTGCCTAAGGATCCCATGT
TCAAGCTGGTTGGTCAGCTGTACAAGATTGTACCCGATATCCTCTTAGAGCAAGGGAAGGCTAAGAACCCTTGGCCCAAC
GTAGACGCTCACAGTGGGGTGCTGCTCCAGTACTACGGCATGAGGGAGATGAATTACTACACAGTCCTGTTCGGAGTGTC
GCGGGCACTGGGCGTTCTGTCCCAGCTCATCTGGAGCAGAGCCCTAGGCTTCCCTCTGGAAAGGCCCAAGTCCATGAGCA
CGGATGCTCTGATGAAGTTTGTGAACTCTGAGTCAGGATAA
ORF - retro_mmus_258 Open Reading Frame is conserved.
Retrocopy - Parental Gene Alignment summary:
| Percent Identity: | 91.81 % |
| Parental protein coverage: | 100. % |
| Number of stop codons detected: | 0 |
| Number of frameshifts detected | 0 |
Retrocopy - Parental Gene Alignment:
| Parental | MALLTAATRLLGAKNSSCLVLAARHASASSTNLKDVLSNLIPKEQARIKTFKQQHGKTVVGQITVDMMYG |
| MALLTAA...LG.KN..CLVLAARHASASSTNLKDVL.NLIPKEQARIKTF...HGKTVVGQITVDMMYG | |
| Retrocopy | MALLTAAAWFLGTKNPPCLVLAARHASASSTNLKDVLRNLIPKEQARIKTFRKKHGKTVVGQITVDMMYG |
| Parental | GMRGMKGLVYETSVLDPDEGIRFRGYSIPECQKMLPKAKGGEEPLPEGLFWLLVTGQMPTEEQVSWLSRE |
| GMRGMKGLVYETSVLDPDEGIRFRGYSIPECQK.LPKAKGG.EPLPEGLFWLLVTGQMPTEEQVSWLS.E | |
| Retrocopy | GMRGMKGLVYETSVLDPDEGIRFRGYSIPECQKLLPKAKGGKEPLPEGLFWLLVTGQMPTEEQVSWLSQE |
| Parental | WAKRAALPSHVVTMLDNFPTNLHPMSQLSAAITALNSESNFARAYAEGMNRAKYWELIYEDCMDLIAKLP |
| W.KRAALPSHVVTMLDNFPT.LHPMSQLSAAIT.LN.ESNFARAYA.GMNR.KYWEL.YEDCMDL.AKLP | |
| Retrocopy | WVKRAALPSHVVTMLDNFPTKLHPMSQLSAAITVLNNESNFARAYAQGMNRTKYWELTYEDCMDLLAKLP |
| Parental | CVAAKIYRNLYREGSSIGAIDSRLDWSHNFTNMLGYTDPQFTELMRLYLTIHSDHEGGNVSAHTSHLVGS |
| CVAAKIYRNLYRE...I.AIDS.LDWSHNFTNMLGYTDPQFTELMRLYLTIHSDHEGGNVSAHTSHLVGS | |
| Retrocopy | CVAAKIYRNLYREDRNIEAIDSKLDWSHNFTNMLGYTDPQFTELMRLYLTIHSDHEGGNVSAHTSHLVGS |
| Parental | ALSDPYLSFAAAMNGLAGPLHGLANQEVLVWLTQLQKEVGKDVSDEKLRDYIWNTLNSGRVVPGYGHAVL |
| ALSDPYLSFAAA.NGLAGPLHGLANQEVLVWLTQLQKEVG.D.SDEKL..YIWNTLNSGRVVPGYGHAVL | |
| Retrocopy | ALSDPYLSFAAALNGLAGPLHGLANQEVLVWLTQLQKEVGEDASDEKLKNYIWNTLNSGRVVPGYGHAVL |
| Parental | RKTDPRYSCQREFALKHLPKDPMFKLVAQLYKIVPNILLEQGKAKNPWPNVDAHSGVLLQYYGMTEMNYY |
| RKTDPRYSCQREFALKHLPKDPMFKLV.QLYKIVP.ILLEQGKAKNPWPNVDAHSGVLLQYYGM.EMNYY | |
| Retrocopy | RKTDPRYSCQREFALKHLPKDPMFKLVGQLYKIVPDILLEQGKAKNPWPNVDAHSGVLLQYYGMREMNYY |
| Parental | TVLFGVSRALGVLAQLIWSRALGFPLERPKSMSTDGLMKFVDSK |
| TVLFGVSRALGVL.QLIWSRALGFPLERPKSMSTD.LMKFV.S. | |
| Retrocopy | TVLFGVSRALGVLSQLIWSRALGFPLERPKSMSTDALMKFVNSE |
Legend:
| * | Stop codon |
| > | Forward frameshift by one nucleotide |
| < | Reverse frameshift by one nucleotide |
(Hint: click retrocopy or parental gene accession number on the plot's legend, to show / hide expression level values)
Expression validation based on RNA-Seq data:
| Library | Retrocopy expression | Parental gene expression |
|---|---|---|
| SRP007412_brain | 0 .19 RPM | 239 .66 RPM |
| SRP007412_cerebellum | 0 .13 RPM | 209 .59 RPM |
| SRP007412_heart | 0 .47 RPM | 1205 .45 RPM |
| SRP007412_kidney | 0 .23 RPM | 434 .64 RPM |
| SRP007412_liver | 0 .05 RPM | 98 .69 RPM |
| SRP007412_testis | 154 .69 RPM | 60 .51 RPM |
RNA Polymerase II actvity near the 5' end of retro_mmus_258 was not detected
No EST(s) were mapped for retro_mmus_258 retrocopy.
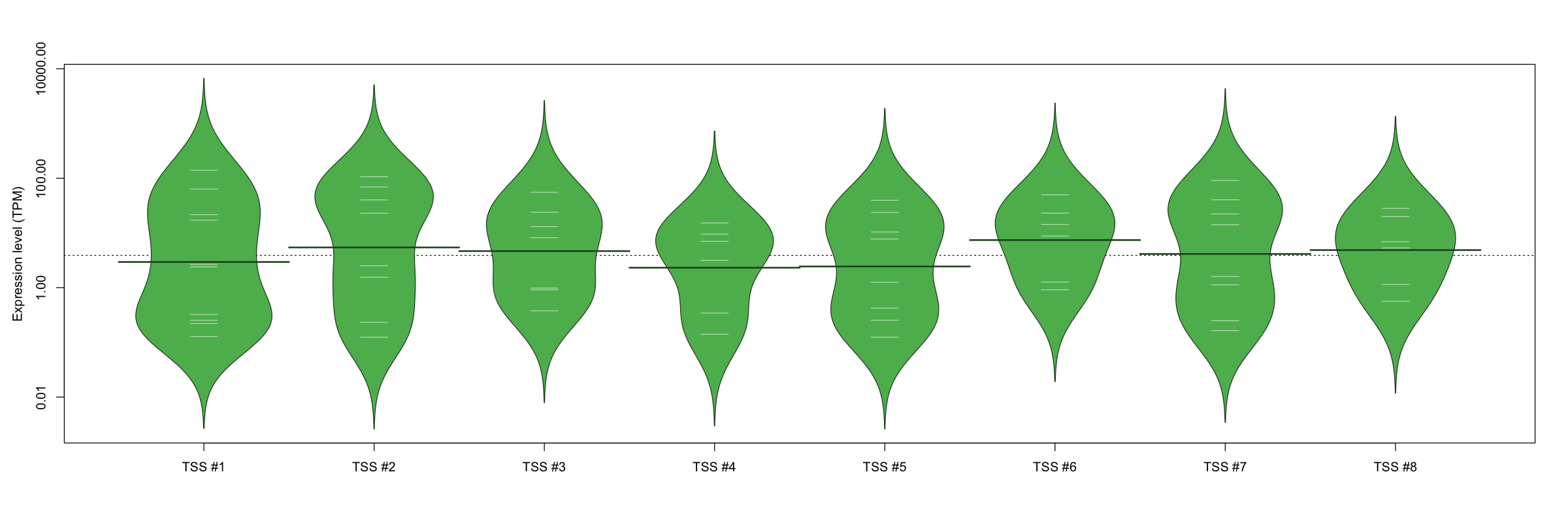
| TSS No. | TSS Name | TSS expression level (Expr) in TPM range: | ||||
|---|---|---|---|---|---|---|
| no expression | 0 < Expr ≤ 1 | 1 < Expr ≤ 5 | 5 < Expr ≤ 10 | Expr > 10 | ||
| TSS #1 | TSS_7761 | 1062 libraries | 4 libraries | 2 libraries | 0 libraries | 4 libraries |
| TSS #2 | TSS_7762 | 1064 libraries | 2 libraries | 2 libraries | 0 libraries | 4 libraries |
| TSS #3 | TSS_7763 | 1065 libraries | 3 libraries | 0 libraries | 1 library | 3 libraries |
| TSS #4 | TSS_7764 | 1066 libraries | 2 libraries | 1 library | 2 libraries | 1 library |
| TSS #5 | TSS_7765 | 1064 libraries | 3 libraries | 1 library | 1 library | 3 libraries |
| TSS #6 | TSS_7766 | 1066 libraries | 1 library | 1 library | 1 library | 3 libraries |
| TSS #7 | TSS_7767 | 1064 libraries | 2 libraries | 2 libraries | 0 libraries | 4 libraries |
| TSS #8 | TSS_7768 | 1066 libraries | 1 library | 1 library | 2 libraries | 2 libraries |
The graphical summary, for retro_mmus_258 TSS expression levels > 0 TPM .
TSS expression levels were studied across 1072 TSS-CAGE libraries, based on FANTOM5 data.
The expression values were visualized using beanplot. If you have any doubts, how to read it, read more in Kampstra P (2008)
retro_mmus_258 was not experimentally validated.
Retrocopy orthology:
Retrocopy retro_mmus_258 has 1 orthologous retrocopies within eutheria group .| Species | RetrogeneDB ID |
|---|---|
| Rattus norvegicus | retro_rnor_2518 |